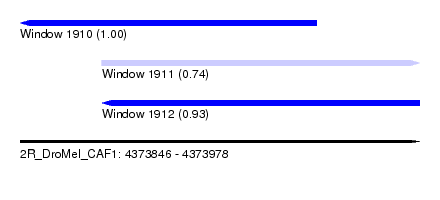
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,373,846 – 4,373,978 |
| Length | 132 |
| Max. P | 0.996814 |

| Location | 4,373,846 – 4,373,944 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 78.17 |
| Mean single sequence MFE | -19.43 |
| Consensus MFE | -17.14 |
| Energy contribution | -16.93 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.75 |
| SVM RNA-class probability | 0.996814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
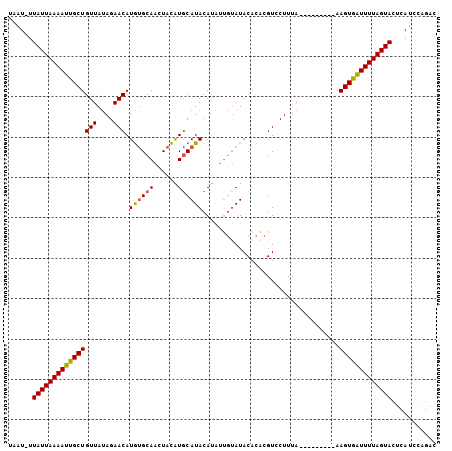
>2R_DroMel_CAF1 4373846 98 - 20766785 UAAAGGACAAGUGUAUGAUUCGUGUAUGUACAUACUUGCAUAUGUUCUGUAACAGCGAUUUUAAUAA-AUUGUAACUCGAUCAAUAAAAAUUGUAUAAC ...((((((.(((((......((((....))))...))))).))))))......((((((((.((..-((((.....))))..)).))))))))..... ( -17.30) >DroSec_CAF1 44521 86 - 1 UAAAGGACGUGUGUAUACAAUAUGUAUGCAUGUAGUUGCACAUGUUCUAUAACAGCAAUUUUAAUAA-AUUAUAACUCGAUCAAUAG------------ ...((((((((((((.((.(((((....))))).))))))))))))))...................-...................------------ ( -20.70) >DroSim_CAF1 44769 87 - 1 UAAAGGACGUGUGUAUACAAUAUGUAUGCAUGUAGUUGCGCAUGUUCUAUAACAGCAAUUUUAAUAAUAUUAUAACUCGAUCAAUAG------------ ...((((((((((((.((.(((((....))))).)))))))))))))).......................................------------ ( -20.30) >consensus UAAAGGACGUGUGUAUACAAUAUGUAUGCAUGUAGUUGCACAUGUUCUAUAACAGCAAUUUUAAUAA_AUUAUAACUCGAUCAAUAG____________ ...((((((((((((.((.(((((....))))).))))))))))))))................................................... (-17.14 = -16.93 + -0.21)

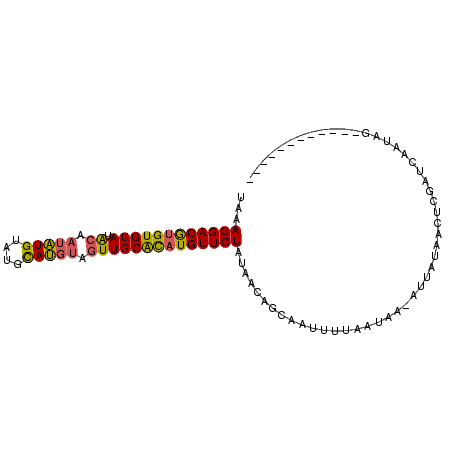
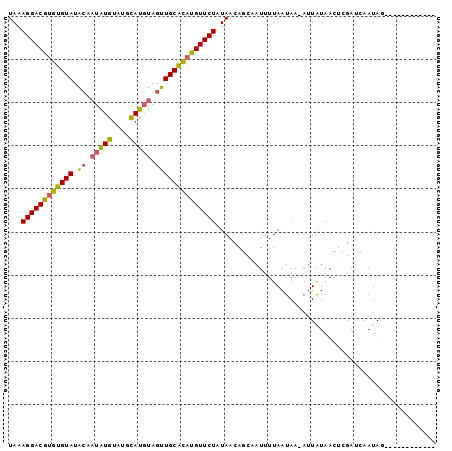
| Location | 4,373,873 – 4,373,978 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 79.22 |
| Mean single sequence MFE | -16.92 |
| Consensus MFE | -12.83 |
| Energy contribution | -12.83 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.742269 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
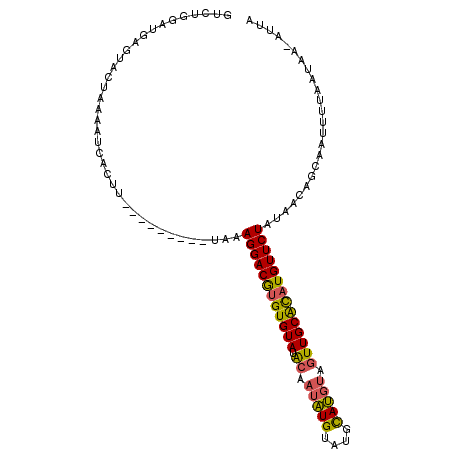
>2R_DroMel_CAF1 4373873 105 + 20766785 CAAU-UUAUUAAAAUCGCUGUUACAGAACAUAUGCAAGUAUGUACAUACACGAAUCAUACACUUGUCCUUUAAAAGCUAUAAAGUGGUUUUAGUACUCAUACGCUC ....-.(((((((((((((.(((.((.......((((((.(((.............))).))))))..........)).))))))))))))))))........... ( -15.95) >DroSec_CAF1 44536 96 + 1 UAAU-UUAUUAAAAUUGCUGUUAUAGAACAUGUGCAACUACAUGCAUACAUAUUGUAUACACACGUCCUUUA---------AAGUGAUUUUAGUACUCAUCCAGAC ....-.(((((((((..((.(((.((.((.((((((......)))((((.....))))..))).)).)).))---------)))..)))))))))........... ( -18.10) >DroSim_CAF1 44784 97 + 1 UAAUAUUAUUAAAAUUGCUGUUAUAGAACAUGCGCAACUACAUGCAUACAUAUUGUAUACACACGUCCUUUA---------AAGUGAUUUUAGUACUCAUCCAGAC ......(((((((((..((.(((.((.((....(((......)))((((.....))))......)).)).))---------)))..)))))))))........... ( -16.70) >consensus UAAU_UUAUUAAAAUUGCUGUUAUAGAACAUGUGCAACUACAUGCAUACAUAUUGUAUACACACGUCCUUUA_________AAGUGAUUUUAGUACUCAUCCAGAC ......((((((((((((((((....))).((((((......))))))..................................)))))))))))))........... (-12.83 = -12.83 + 0.01)

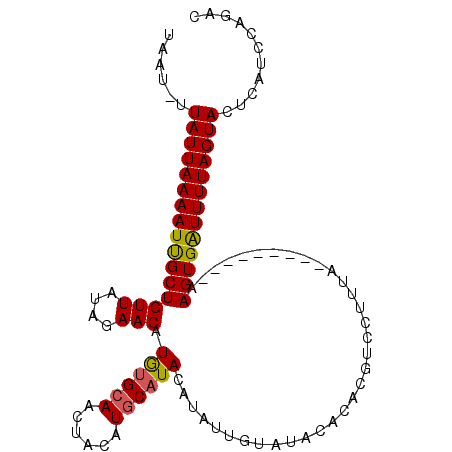
| Location | 4,373,873 – 4,373,978 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 79.22 |
| Mean single sequence MFE | -22.17 |
| Consensus MFE | -17.14 |
| Energy contribution | -16.93 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932346 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
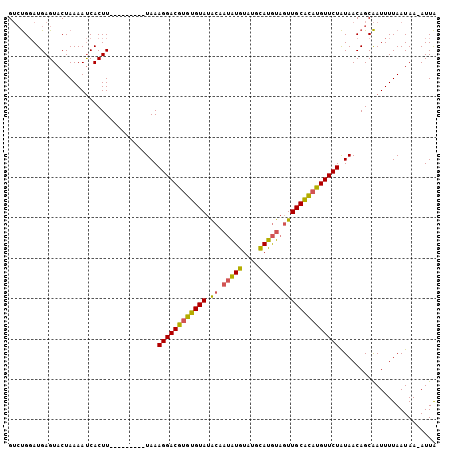
>2R_DroMel_CAF1 4373873 105 - 20766785 GAGCGUAUGAGUACUAAAACCACUUUAUAGCUUUUAAAGGACAAGUGUAUGAUUCGUGUAUGUACAUACUUGCAUAUGUUCUGUAACAGCGAUUUUAAUAA-AUUG (((((((((.............(((((.......)))))..(((((((...((........))..))))))))))))))))........((((((....))-)))) ( -18.50) >DroSec_CAF1 44536 96 - 1 GUCUGGAUGAGUACUAAAAUCACUU---------UAAAGGACGUGUGUAUACAAUAUGUAUGCAUGUAGUUGCACAUGUUCUAUAACAGCAAUUUUAAUAA-AUUA .....(((...((.((((((..(((---------((.((((((((((((.((.(((((....))))).)))))))))))))).))).))..)))))).)).-))). ( -24.30) >DroSim_CAF1 44784 97 - 1 GUCUGGAUGAGUACUAAAAUCACUU---------UAAAGGACGUGUGUAUACAAUAUGUAUGCAUGUAGUUGCGCAUGUUCUAUAACAGCAAUUUUAAUAAUAUUA .....((((..((.((((((..(((---------((.((((((((((((.((.(((((....))))).)))))))))))))).))).))..)))))).)).)))). ( -23.70) >consensus GUCUGGAUGAGUACUAAAAUCACUU_________UAAAGGACGUGUGUAUACAAUAUGUAUGCAUGUAGUUGCACAUGUUCUAUAACAGCAAUUUUAAUAA_AUUA .....................................((((((((((((.((.(((((....))))).))))))))))))))........................ (-17.14 = -16.93 + -0.21)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:59 2006