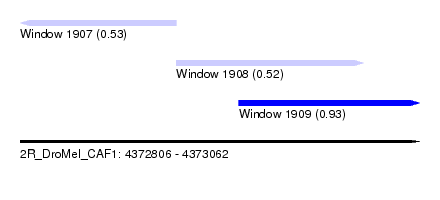
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,372,806 – 4,373,062 |
| Length | 256 |
| Max. P | 0.932093 |

| Location | 4,372,806 – 4,372,906 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 86.78 |
| Mean single sequence MFE | -42.32 |
| Consensus MFE | -28.74 |
| Energy contribution | -29.30 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.527845 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4372806 100 - 20766785 CGAACCGAGGGACUCGGG-----GACAAGGCGGCCAUCGGUAAUCCAAGGGUCCGAUGGCACG---------CAAUCCC-GUCUCCACCUUCGCCAUUUUGCUGAGCCGGCAGCA .....((((((....(((-----(((...(((((((((((..(((....))))))))))).))---------)......-)))))).))))))......(((((......))))) ( -42.40) >DroSec_CAF1 43472 96 - 1 ----CCGAUGGACUCGGG-----GACAAGGCGGCCAUCGGUAAUCCAAGGGUCCGAUGGCACG---------CAAUCCC-GCCUCCACCUCCGCCAUUUUGCUGAACCGGCAGCA ----..(.((((..((((-----(.....(((((((((((..(((....))))))))))).))---------)..))))-)..)))).)..........(((((......))))) ( -39.30) >DroSim_CAF1 43705 100 - 1 CGAACCGAGGGACUCGGG-----GACAAGGCGGCCAUCGGUAAUCCAAGGGUCCGAUGGCACG---------CAAUCCC-GCCUCCACCUCCGCCAUUUUGCUGAACCGGCAGCA ......((((((..((((-----(.....(((((((((((..(((....))))))))))).))---------)..))))-)..))..))))........(((((......))))) ( -39.60) >DroEre_CAF1 39972 109 - 1 CGAGCCGAGGGACUCGGG-----GACAAGGCGGCCAUCGGUAAUCCAAGGGUCCGAUGGCACACCCCGGGCACAAUCCCGCCCUCCUGCUCCGCCAU-UUGCUGAACCGGCAGCA .((((.(((((....(((-----(.(.....)((((((((..(((....)))))))))))...))))(((......))).)))))..))))......-.(((((......))))) ( -46.80) >DroYak_CAF1 41880 104 - 1 UGAAGCGAGGGACUCGGGGCAGGGGUAAGGCGGCCAUCAGUAAUCCAAGGGUCCGAUGGCACG---------CAAUCCC-CCCUCCUCCUCCGCCAU-UUGCUGAACCGGCAGCA ....(((..(((...((((..((((....(((((((((.(..(((....)))).)))))).))---------)...)))-).)))))))..)))...-.(((((......))))) ( -43.50) >consensus CGAACCGAGGGACUCGGG_____GACAAGGCGGCCAUCGGUAAUCCAAGGGUCCGAUGGCACG_________CAAUCCC_GCCUCCACCUCCGCCAUUUUGCUGAACCGGCAGCA ....(((((...)))))...........((((((((((((..(((....)))))))))))....................(......)...))))....(((((......))))) (-28.74 = -29.30 + 0.56)

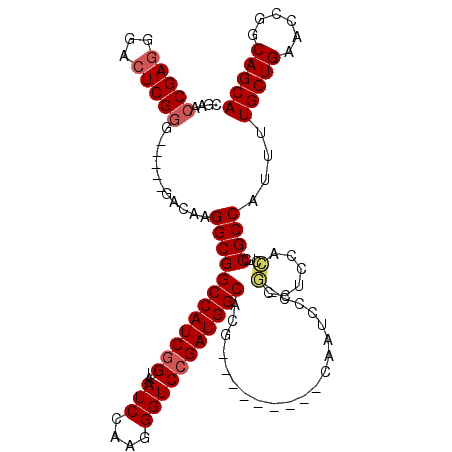
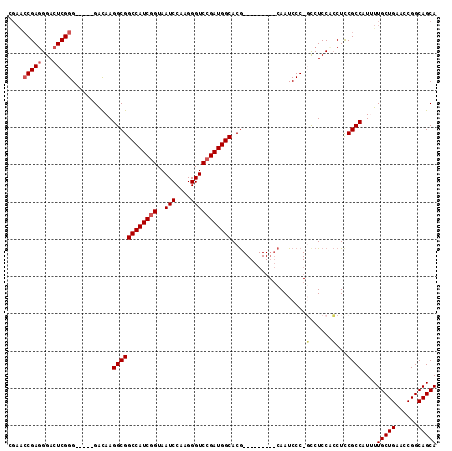
| Location | 4,372,906 – 4,373,026 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.00 |
| Mean single sequence MFE | -31.30 |
| Consensus MFE | -25.70 |
| Energy contribution | -25.38 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.82 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.516618 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4372906 120 + 20766785 CAGACUGACUGCCGCCAGCGAAAUCAGUAAAUUUAUGAGAAAAGUCCUUGCCCAAGGAGAAACACACCCGGCGAACCAGAGAUCAAUUAAUUAGAACUCGUCGCCGGGACUCUCGGGAAA ...(((((.(..((....)).).)))))...(((.(((((....(((((....)))))........((((((((....(((.((.........)).))).))))))))..))))).))). ( -36.70) >DroSec_CAF1 43568 102 + 1 ------------------UGAAAUCAGUAAAGAUAUGAGAAAAGUCCUUGCCCAAGGAGAAACGCACCCGGCGAACCAGAGAGCAAUUAAUUAGAACUCGUCGCCGGGACUCUCGGGAAA ------------------....(((......))).(((((....(((((....)))))........((((((((....(((..(.........)..))).))))))))..)))))..... ( -29.40) >DroSim_CAF1 43805 120 + 1 CAGACUGACUGCCGCCAGUGAAAUCAGUAAAGAUAUGAGAAAAGUCCUUGCCCAAGGAGAAACGCAACCGGCGAACCAGAGAGCAAUUAAUUAGAACUCGUCGCCGGUACUCUCGGGAAA ...((((((((....))).....))))).......(((((....(((((....)))))........((((((((....(((..(.........)..))).))))))))..)))))..... ( -31.30) >DroEre_CAF1 40081 120 + 1 CAGACUGACUGCCGCCAGUGAAAUCAGUAAAAAUAUGAGAAAAGUCCUUGCCCAAGGAGAGACCCACCCGGCGAGCCAGAAAGCAAUCAAUUAGAACUCGUCGUCGGGACUCUCGGGAAA ..((((.((((....))))....(((.........)))....))))....(((...(((((.(((...(((((((.....................)))))))..))).))))))))... ( -29.80) >DroYak_CAF1 41984 120 + 1 CAGACUGACUGCCGCCAGUGAAAUCUGUAAAAAUAUGAGAAUAGUCCUUGCCCAGGGAGAAACCCACCCGGCGAACCAGAAAGCAAUUAAUUAGAACUCGUCGUCGGGACUCUCGGGAAA ..(((((((((....))))....((((((....))).))).)))))....(((.(((.....))).((((((((....((...(.........)...)).))))))))......)))... ( -29.30) >consensus CAGACUGACUGCCGCCAGUGAAAUCAGUAAAAAUAUGAGAAAAGUCCUUGCCCAAGGAGAAACCCACCCGGCGAACCAGAGAGCAAUUAAUUAGAACUCGUCGCCGGGACUCUCGGGAAA ...................................(((((....(((((....)))))........((((((((....(((...............))).))))))))..)))))..... (-25.70 = -25.38 + -0.32)

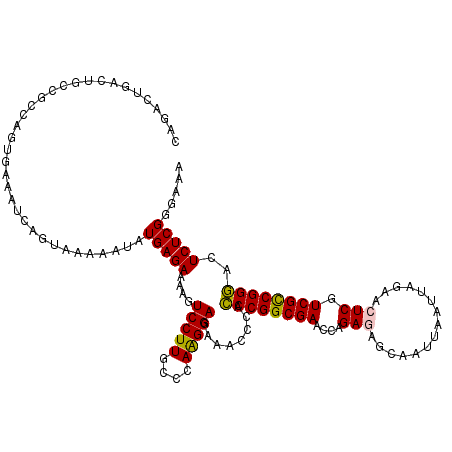
| Location | 4,372,946 – 4,373,062 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.84 |
| Mean single sequence MFE | -35.38 |
| Consensus MFE | -30.78 |
| Energy contribution | -30.66 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932093 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
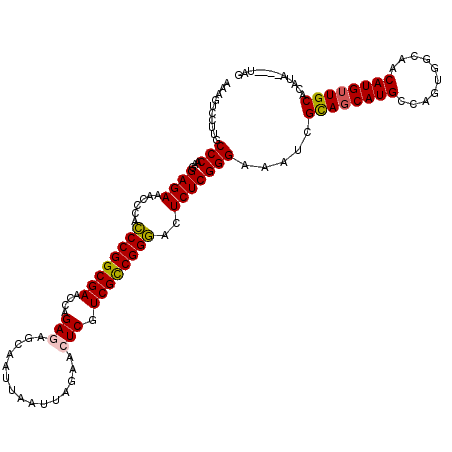
>2R_DroMel_CAF1 4372946 116 + 20766785 AAAGUCCUUGCCCAAGGAGAAACACACCCGGCGAACCAGAGAUCAAUUAAUUAGAACUCGUCGCCGGGACUCUCGGGAAAUCGCAGCAUGUCAGCGGCAACAUGUUGCACAUA----UAG ..........(((...((((......((((((((....(((.((.........)).))).))))))))..))))))).....(((((((((........))))))))).....----... ( -38.90) >DroSec_CAF1 43590 116 + 1 AAAGUCCUUGCCCAAGGAGAAACGCACCCGGCGAACCAGAGAGCAAUUAAUUAGAACUCGUCGCCGGGACUCUCGGGAAAUCGUAGCAUGCCAGUGGCAACAUGUUGCACAUA----UAG ..........(((...((((......((((((((....(((..(.........)..))).))))))))..))))))).....((((((((((...))))...)))))).....----... ( -34.40) >DroSim_CAF1 43845 116 + 1 AAAGUCCUUGCCCAAGGAGAAACGCAACCGGCGAACCAGAGAGCAAUUAAUUAGAACUCGUCGCCGGUACUCUCGGGAAAUCGCAGCAUGCCAGUGGCAACAUGUUGCACAUA----UAG ..........(((...((((......((((((((....(((..(.........)..))).))))))))..))))))).....((((((((((...))))...)))))).....----... ( -34.60) >DroEre_CAF1 40121 116 + 1 AAAGUCCUUGCCCAAGGAGAGACCCACCCGGCGAGCCAGAAAGCAAUCAAUUAGAACUCGUCGUCGGGACUCUCGGGAAAUCGCAUCAUGCGAGUGGCAACAUGUUGCACAUA----UAU ..........(((...(((((.(((...(((((((.....................)))))))..))).))))))))...((((.....))))(((((((....)))).))).----... ( -35.10) >DroYak_CAF1 42024 120 + 1 AUAGUCCUUGCCCAGGGAGAAACCCACCCGGCGAACCAGAAAGCAAUUAAUUAGAACUCGUCGUCGGGACUCUCGGGAAAUCGCAGCAUGCGAGUGGCAACAUGUUGCACAUAUGUAUAU ...((.....(((.(((.....))).((((((((....((...(.........)...)).))))))))......))).....))...(((((.(((((((....)))).))).))))).. ( -33.90) >consensus AAAGUCCUUGCCCAAGGAGAAACCCACCCGGCGAACCAGAGAGCAAUUAAUUAGAACUCGUCGCCGGGACUCUCGGGAAAUCGCAGCAUGCCAGUGGCAACAUGUUGCACAUA____UAG ..........(((...((((......((((((((....(((...............))).))))))))..))))))).....((((((((..........))))))))............ (-30.78 = -30.66 + -0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:57 2006