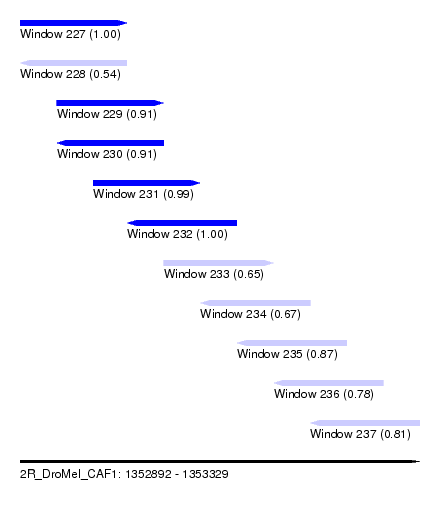
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,352,892 – 1,353,329 |
| Length | 437 |
| Max. P | 0.998311 |

| Location | 1,352,892 – 1,353,009 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.94 |
| Mean single sequence MFE | -29.16 |
| Consensus MFE | -26.33 |
| Energy contribution | -26.53 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.89 |
| SVM RNA-class probability | 0.997575 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1352892 117 + 20766785 CCUCCGGCAUGACAUUUACUCUCCGAAUUACUAUAAACUGCUGCAUUUGAACGAAAGAAGCAAAGUGGCAGCAAAUAACGACUAACUGCCACCCCGGAGAUCGCCGCCCG---CCCACCC ....((((..((.......((((((.............((((.(.(((....))).).))))..(((((((..............)))))))..))))))))))))....---....... ( -27.25) >DroSec_CAF1 71019 120 + 1 CCUCCGGCAUGACAUUUACUCUCCGAAUUACUAUAAACUGCUGCAUUUGAACGAAAGAAGCAAAGUGGCAGCAAAUAACAACUAACUGCCACCCCGGAGAUCGCCGCCCGCCGCCCAUCC .....(((...........((((((.............((((.(.(((....))).).))))..(((((((..............)))))))..))))))..((.....)).)))..... ( -28.64) >DroSim_CAF1 49187 120 + 1 CCUCCGGCAUGACAUUUACUCUCCGAAUUACUAUAAACUGCUGCAUUUGAACGAAAGAAGCAAAGUGGCAGCAAAUAACAACUAACUGCCACCCCGGAGAUCGCCGCCCGCCGCCCAUCC .....(((...........((((((.............((((.(.(((....))).).))))..(((((((..............)))))))..))))))..((.....)).)))..... ( -28.64) >DroEre_CAF1 51651 113 + 1 CCUCCGGCAUGACAUUUACUCUCCGAAUUACUAUAAACUGCUGCAUUUGAACGAAAGAAGCAAAGUGGCAGCAAAUAACGGCUAACUGCCACCCCGGAGAUCGCCGCCCAC---CC---- ....((((..((.......((((((.............((((((.((((..(....)...))))...))))))......(((.....)))....)))))))))))).....---..---- ( -32.91) >DroYak_CAF1 38911 113 + 1 CCUCAGGCAGGACAUUUACUCUCCGAAUUACUAUAAACUGCUGCAUUUGAACGAAAGAAGCAAAGUGGCAGCAAAUAACAACUAACUGCCACCCCGGAGAUCGCCGCCCAC---CC---- .....(((.((........((((((.............((((.(.(((....))).).))))..(((((((..............)))))))..))))))...)))))...---..---- ( -28.34) >consensus CCUCCGGCAUGACAUUUACUCUCCGAAUUACUAUAAACUGCUGCAUUUGAACGAAAGAAGCAAAGUGGCAGCAAAUAACAACUAACUGCCACCCCGGAGAUCGCCGCCCGC__CCCA_CC ....((((..((.......((((((.............((((.(.(((....))).).))))..(((((((..............)))))))..)))))))))))).............. (-26.33 = -26.53 + 0.20)

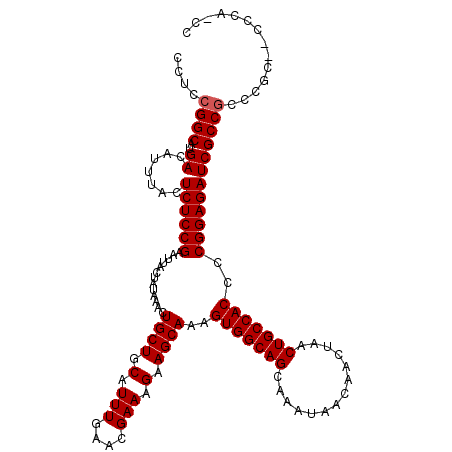
| Location | 1,352,892 – 1,353,009 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.94 |
| Mean single sequence MFE | -39.50 |
| Consensus MFE | -34.20 |
| Energy contribution | -34.16 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.537836 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1352892 117 - 20766785 GGGUGGG---CGGGCGGCGAUCUCCGGGGUGGCAGUUAGUCGUUAUUUGCUGCCACUUUGCUUCUUUCGUUCAAAUGCAGCAGUUUAUAGUAAUUCGGAGAGUAAAUGUCAUGCCGGAGG .....((---((((((....((((((((((((((((.(((....))).)))))))))(((((.(............).)))))...........))))))).....)))).))))..... ( -38.60) >DroSec_CAF1 71019 120 - 1 GGAUGGGCGGCGGGCGGCGAUCUCCGGGGUGGCAGUUAGUUGUUAUUUGCUGCCACUUUGCUUCUUUCGUUCAAAUGCAGCAGUUUAUAGUAAUUCGGAGAGUAAAUGUCAUGCCGGAGG .......(((((((((....((((((((((((((((.(((....))).)))))))))(((((.(............).)))))...........))))))).....)))).))))).... ( -41.60) >DroSim_CAF1 49187 120 - 1 GGAUGGGCGGCGGGCGGCGAUCUCCGGGGUGGCAGUUAGUUGUUAUUUGCUGCCACUUUGCUUCUUUCGUUCAAAUGCAGCAGUUUAUAGUAAUUCGGAGAGUAAAUGUCAUGCCGGAGG .......(((((((((....((((((((((((((((.(((....))).)))))))))(((((.(............).)))))...........))))))).....)))).))))).... ( -41.60) >DroEre_CAF1 51651 113 - 1 ----GG---GUGGGCGGCGAUCUCCGGGGUGGCAGUUAGCCGUUAUUUGCUGCCACUUUGCUUCUUUCGUUCAAAUGCAGCAGUUUAUAGUAAUUCGGAGAGUAAAUGUCAUGCCGGAGG ----.(---(((((((....((((((((((((((((............)))))))))(((((.(............).)))))...........))))))).....)))).))))..... ( -35.40) >DroYak_CAF1 38911 113 - 1 ----GG---GUGGGCGGCGAUCUCCGGGGUGGCAGUUAGUUGUUAUUUGCUGCCACUUUGCUUCUUUCGUUCAAAUGCAGCAGUUUAUAGUAAUUCGGAGAGUAAAUGUCCUGCCUGAGG ----((---(((((((....((((((((((((((((.(((....))).)))))))))(((((.(............).)))))...........))))))).....))))).)))).... ( -40.30) >consensus GG_UGGG__GCGGGCGGCGAUCUCCGGGGUGGCAGUUAGUUGUUAUUUGCUGCCACUUUGCUUCUUUCGUUCAAAUGCAGCAGUUUAUAGUAAUUCGGAGAGUAAAUGUCAUGCCGGAGG ..........(.(((((((.((((((((((((((((.(((....))).)))))))))(((((.(............).)))))...........))))))).....))))..))).)... (-34.20 = -34.16 + -0.04)

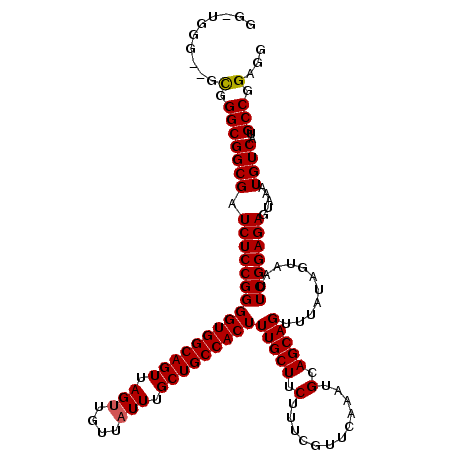
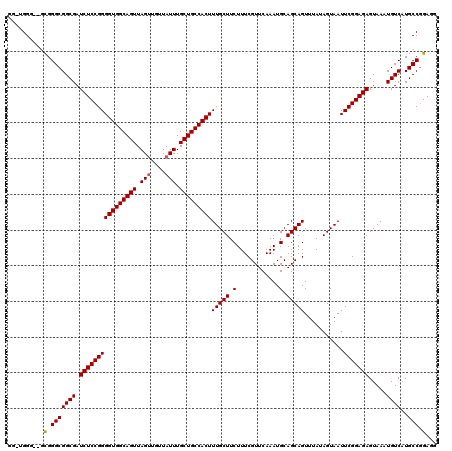
| Location | 1,352,932 – 1,353,049 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.67 |
| Mean single sequence MFE | -32.18 |
| Consensus MFE | -28.30 |
| Energy contribution | -28.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.907494 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1352932 117 + 20766785 CUGCAUUUGAACGAAAGAAGCAAAGUGGCAGCAAAUAACGACUAACUGCCACCCCGGAGAUCGCCGCCCG---CCCACCCGGCGCAUUAUCAAUUUAAACGAAUGGCACACGAGUGUGCC ...((((((..(....)..((...(((((((..............))))))).((((.(...((.....)---)...))))).))..............))))))(((((....))))). ( -32.04) >DroSec_CAF1 71059 120 + 1 CUGCAUUUGAACGAAAGAAGCAAAGUGGCAGCAAAUAACAACUAACUGCCACCCCGGAGAUCGCCGCCCGCCGCCCAUCCGGCGCAUUAUCAAUUUAAACGAAUGGCACACGAGUGUGCC ...((((((..(....)..((...(((((((..............))))))).(((((....((.(....).))...))))).))..............))))))(((((....))))). ( -35.74) >DroSim_CAF1 49227 120 + 1 CUGCAUUUGAACGAAAGAAGCAAAGUGGCAGCAAAUAACAACUAACUGCCACCCCGGAGAUCGCCGCCCGCCGCCCAUCCGGCGCAUUAUCAAUUUAAACGAAUGGCACACGAGUGUGCC ...((((((..(....)..((...(((((((..............))))))).(((((....((.(....).))...))))).))..............))))))(((((....))))). ( -35.74) >DroEre_CAF1 51691 111 + 1 CUGCAUUUGAACGAAAGAAGCAAAGUGGCAGCAAAUAACGGCUAACUGCCACCCCGGAGAUCGCCGCCCAC---CC------AGCAUUAUCAAUUUAAGCGAAUGGCACACGAGUGUGCC .(((..((((.(....)..((...(((((((..............)))))))..(((......))).....---..------.))....)))).....)))...((((((....)))))) ( -28.44) >DroYak_CAF1 38951 111 + 1 CUGCAUUUGAACGAAAGAAGCAAAGUGGCAGCAAAUAACAACUAACUGCCACCCCGGAGAUCGCCGCCCAC---CC------UGCAUUAUCAAUUUAAACGAAUGGCACACGAGUGUGCC ...((((((..(....)..(((..(((((((..............)))))))..(((......))).....---..------)))..............))))))(((((....))))). ( -28.94) >consensus CUGCAUUUGAACGAAAGAAGCAAAGUGGCAGCAAAUAACAACUAACUGCCACCCCGGAGAUCGCCGCCCGC__CCCA_CCGGCGCAUUAUCAAUUUAAACGAAUGGCACACGAGUGUGCC ...((((((..(....)..((...(((((((..............)))))))..(((......))).................))..............))))))(((((....))))). (-28.30 = -28.30 + 0.00)

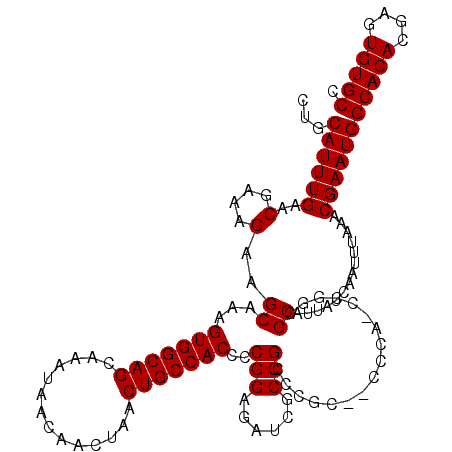
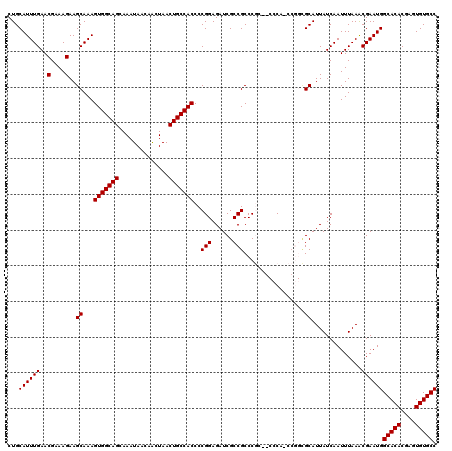
| Location | 1,352,932 – 1,353,049 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.67 |
| Mean single sequence MFE | -42.74 |
| Consensus MFE | -34.32 |
| Energy contribution | -35.32 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908931 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1352932 117 - 20766785 GGCACACUCGUGUGCCAUUCGUUUAAAUUGAUAAUGCGCCGGGUGGG---CGGGCGGCGAUCUCCGGGGUGGCAGUUAGUCGUUAUUUGCUGCCACUUUGCUUCUUUCGUUCAAAUGCAG ((((((....))))))..................((((((.....))---)((((((.((....((((((((((((.(((....))).))))))))))))..))..))))))....))). ( -42.30) >DroSec_CAF1 71059 120 - 1 GGCACACUCGUGUGCCAUUCGUUUAAAUUGAUAAUGCGCCGGAUGGGCGGCGGGCGGCGAUCUCCGGGGUGGCAGUUAGUUGUUAUUUGCUGCCACUUUGCUUCUUUCGUUCAAAUGCAG ((((((....))))))..................(((((((......))))((((((.((....((((((((((((.(((....))).))))))))))))..))..))))))....))). ( -45.50) >DroSim_CAF1 49227 120 - 1 GGCACACUCGUGUGCCAUUCGUUUAAAUUGAUAAUGCGCCGGAUGGGCGGCGGGCGGCGAUCUCCGGGGUGGCAGUUAGUUGUUAUUUGCUGCCACUUUGCUUCUUUCGUUCAAAUGCAG ((((((....))))))..................(((((((......))))((((((.((....((((((((((((.(((....))).))))))))))))..))..))))))....))). ( -45.50) >DroEre_CAF1 51691 111 - 1 GGCACACUCGUGUGCCAUUCGCUUAAAUUGAUAAUGCU------GG---GUGGGCGGCGAUCUCCGGGGUGGCAGUUAGCCGUUAUUUGCUGCCACUUUGCUUCUUUCGUUCAAAUGCAG ((((((....))))))..................(((.------..---.(((((((.((....((((((((((((............))))))))))))..))..)))))))...))). ( -38.40) >DroYak_CAF1 38951 111 - 1 GGCACACUCGUGUGCCAUUCGUUUAAAUUGAUAAUGCA------GG---GUGGGCGGCGAUCUCCGGGGUGGCAGUUAGUUGUUAUUUGCUGCCACUUUGCUUCUUUCGUUCAAAUGCAG ((((((....))))))..................((((------..---.(((((((.((....((((((((((((.(((....))).))))))))))))..))..)))))))..)))). ( -42.00) >consensus GGCACACUCGUGUGCCAUUCGUUUAAAUUGAUAAUGCGCCGG_UGGG__GCGGGCGGCGAUCUCCGGGGUGGCAGUUAGUUGUUAUUUGCUGCCACUUUGCUUCUUUCGUUCAAAUGCAG ((((((....))))))..................(((.((....)).....((((((.((....((((((((((((.(((....))).))))))))))))..))..))))))....))). (-34.32 = -35.32 + 1.00)

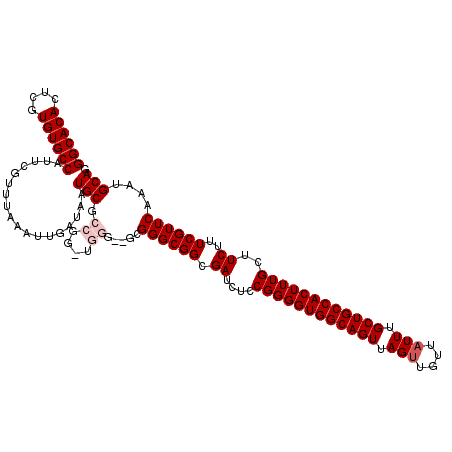
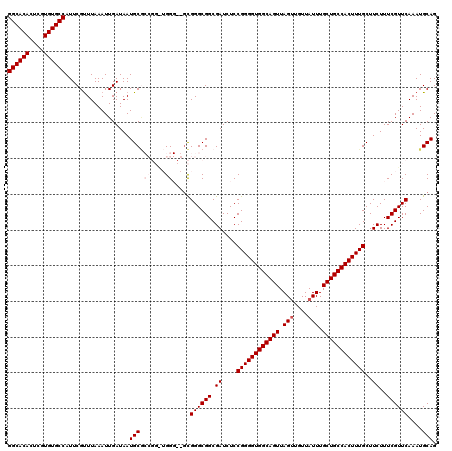
| Location | 1,352,972 – 1,353,089 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.18 |
| Mean single sequence MFE | -35.00 |
| Consensus MFE | -30.00 |
| Energy contribution | -30.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.992120 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1352972 117 + 20766785 ACUAACUGCCACCCCGGAGAUCGCCGCCCG---CCCACCCGGCGCAUUAUCAAUUUAAACGAAUGGCACACGAGUGUGCCAUAAAUUCGGGCUGCAUGACAUUUGCUAAUGUUUGUCCAU ...............(((.(..((.(((((---(((....)).)).................((((((((....))))))))......)))).))..((((((....))))))).))).. ( -35.60) >DroSec_CAF1 71099 120 + 1 ACUAACUGCCACCCCGGAGAUCGCCGCCCGCCGCCCAUCCGGCGCAUUAUCAAUUUAAACGAAUGGCACACGAGUGUGCCAUAAAUUCGGGCUGCAUGACAUUUGCUAAUGUUUGUCCAU ...............(((.(..((.((((((((......))))...................((((((((....))))))))......)))).))..((((((....))))))).))).. ( -37.90) >DroSim_CAF1 49267 120 + 1 ACUAACUGCCACCCCGGAGAUCGCCGCCCGCCGCCCAUCCGGCGCAUUAUCAAUUUAAACGAAUGGCACACGAGUGUGCCAUAAAUUCGGGCUGCAUGACAUUUGCUAAUGUUUGUCCAU ...............(((.(..((.((((((((......))))...................((((((((....))))))))......)))).))..((((((....))))))).))).. ( -37.90) >DroEre_CAF1 51731 111 + 1 GCUAACUGCCACCCCGGAGAUCGCCGCCCAC---CC------AGCAUUAUCAAUUUAAGCGAAUGGCACACGAGUGUGCCAUAAAUUCGGGCUGCAUGACAUUUGCUAAUGUUUGUCCAU ((.....))......(((.(..((.((((..---..------.((.(((......)))))..((((((((....))))))))......)))).))..((((((....))))))).))).. ( -33.60) >DroYak_CAF1 38991 111 + 1 ACUAACUGCCACCCCGGAGAUCGCCGCCCAC---CC------UGCAUUAUCAAUUUAAACGAAUGGCACACGAGUGUGCCAUAAAUUCGGGCUGCAUGACAUUUGCUAAUGUUUGUCCAU ...............(((.(..((.((((..---..------....(((......)))....((((((((....))))))))......)))).))..((((((....))))))).))).. ( -30.00) >consensus ACUAACUGCCACCCCGGAGAUCGCCGCCCGC__CCCA_CCGGCGCAUUAUCAAUUUAAACGAAUGGCACACGAGUGUGCCAUAAAUUCGGGCUGCAUGACAUUUGCUAAUGUUUGUCCAU ...............(((.(..((.((((.................(((......)))....((((((((....))))))))......)))).))..((((((....))))))).))).. (-30.00 = -30.00 + 0.00)

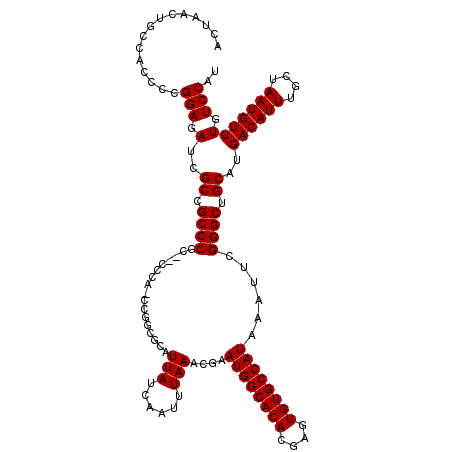
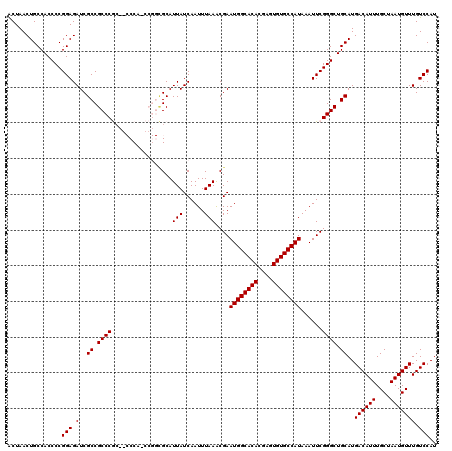
| Location | 1,353,009 – 1,353,129 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.25 |
| Mean single sequence MFE | -32.68 |
| Consensus MFE | -31.68 |
| Energy contribution | -31.68 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.97 |
| SVM decision value | 3.06 |
| SVM RNA-class probability | 0.998311 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1353009 120 - 20766785 CGCAAGUUUGCCAUGGUAUUUAAUUAACGCCGCCGCUCUCAUGGACAAACAUUAGCAAAUGUCAUGCAGCCCGAAUUUAUGGCACACUCGUGUGCCAUUCGUUUAAAUUGAUAAUGCGCC .((..((((((((((((.........))((....))...))))).)))))....(((..(((((.......(((....((((((((....))))))))))).......))))).))))). ( -33.04) >DroSec_CAF1 71139 120 - 1 CGCAAGUUUGCCAUGGUAUUUAAUUAACGCCGCUGCUCUCAUGGACAAACAUUAGCAAAUGUCAUGCAGCCCGAAUUUAUGGCACACUCGUGUGCCAUUCGUUUAAAUUGAUAAUGCGCC .((..((((((((((((.........))((....))...))))).)))))....(((..(((((.......(((....((((((((....))))))))))).......))))).))))). ( -32.94) >DroSim_CAF1 49307 120 - 1 CGCAAGUUUGCCAUGGUAUUUAAUUAACGCCGCUGCUCUCAUGGACAAACAUUAGCAAAUGUCAUGCAGCCCGAAUUUAUGGCACACUCGUGUGCCAUUCGUUUAAAUUGAUAAUGCGCC .((..((((((((((((.........))((....))...))))).)))))....(((..(((((.......(((....((((((((....))))))))))).......))))).))))). ( -32.94) >DroEre_CAF1 51764 118 - 1 CGCAAGUUUGCCAUGGUAUUUAAUUAACGCCGCCGCGCUCAUGGACAAACAUUAGCAAAUGUCAUGCAGCCCGAAUUUAUGGCACACUCGUGUGCCAUUCGCUUAAAUUGAUAAUGCU-- .(((.(((((((((((((......)).(((....))).)))))).))))).........(((((.......(((....((((((((....))))))))))).......))))).))).-- ( -32.84) >DroYak_CAF1 39024 118 - 1 CGCAAGUUUGCCAUGGUAUUUAAUUAACGCCGCUGCUCUCAUGGACAAACAUUAGCAAAUGUCAUGCAGCCCGAAUUUAUGGCACACUCGUGUGCCAUUCGUUUAAAUUGAUAAUGCA-- .(((.((((((((((((.........))((....))...))))).))))).........(((((.......(((....((((((((....))))))))))).......))))).))).-- ( -31.64) >consensus CGCAAGUUUGCCAUGGUAUUUAAUUAACGCCGCUGCUCUCAUGGACAAACAUUAGCAAAUGUCAUGCAGCCCGAAUUUAUGGCACACUCGUGUGCCAUUCGUUUAAAUUGAUAAUGCGCC .(((.((((((((((((.........))((....))...))))).))))).........(((((.......(((....((((((((....))))))))))).......))))).)))... (-31.68 = -31.68 + -0.00)

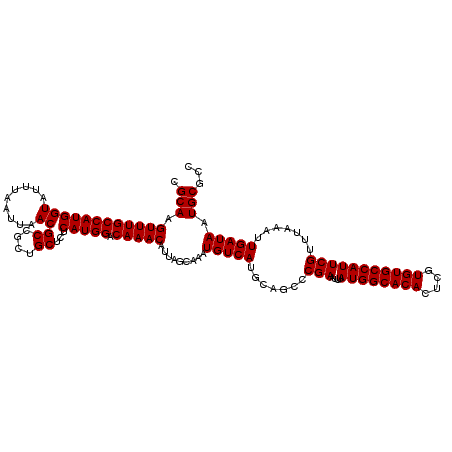
| Location | 1,353,049 – 1,353,169 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -36.10 |
| Consensus MFE | -33.56 |
| Energy contribution | -33.52 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.650248 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1353049 120 + 20766785 AUAAAUUCGGGCUGCAUGACAUUUGCUAAUGUUUGUCCAUGAGAGCGGCGGCGUUAAUUAAAUACCAUGGCAAACUUGCGGCCUGCAGCGCGAAAUCAGAAAUUCUGAAUCGGCGAGCUC ........((((((((.....(((((((.(((((........)))))..((..((.....))..)).)))))))..))))))))((.((.(((..((((.....)))).)))))..)).. ( -34.00) >DroSec_CAF1 71179 119 + 1 AUAAAUUCGGGCUGCAUGACAUUUGCUAAUGUUUGUCCAUGAGAGCAGCGGCGUUAAUUAAAUACCAUGGCAAACUUGCGGCCUGCAGCGCGA-AUCAGAAAUUCUGAAUCGGCGAGCUC ........((((((((.....(((((((.(((((........)))))..((..((.....))..)).)))))))..))))))))((.((.(((-.((((.....)))).)))))..)).. ( -36.90) >DroSim_CAF1 49347 119 + 1 AUAAAUUCGGGCUGCAUGACAUUUGCUAAUGUUUGUCCAUGAGAGCAGCGGCGUUAAUUAAAUACCAUGGCAAACUUGCGGCCUGCAGCGCGA-AUCAGAAAUUCUGAAUCGGCGAGCUC ........((((((((.....(((((((.(((((........)))))..((..((.....))..)).)))))))..))))))))((.((.(((-.((((.....)))).)))))..)).. ( -36.90) >DroEre_CAF1 51802 120 + 1 AUAAAUUCGGGCUGCAUGACAUUUGCUAAUGUUUGUCCAUGAGCGCGGCGGCGUUAAUUAAAUACCAUGGCAAACUUGCGGCCUGCAGCGCGAAAUCAGAAAUUCUGAAUCGGCGGGCUC ........((((((((.((((((....)))))(((.((((((((((....)))))..........)))))))).).))))))))((.((.(((..((((.....)))).)))))..)).. ( -38.00) >DroYak_CAF1 39062 120 + 1 AUAAAUUCGGGCUGCAUGACAUUUGCUAAUGUUUGUCCAUGAGAGCAGCGGCGUUAAUUAAAUACCAUGGCAAACUUGCGGCCUGCAGCGCGAAAUCAGAAAUUCUGAAUCGGCGAGCUC ........((((((((.....(((((((.(((((........)))))..((..((.....))..)).)))))))..))))))))((.((.(((..((((.....)))).)))))..)).. ( -34.70) >consensus AUAAAUUCGGGCUGCAUGACAUUUGCUAAUGUUUGUCCAUGAGAGCAGCGGCGUUAAUUAAAUACCAUGGCAAACUUGCGGCCUGCAGCGCGAAAUCAGAAAUUCUGAAUCGGCGAGCUC ........((((((((.....(((((((.(((((........)))))..((..((.....))..)).)))))))..))))))))((.((.(((..((((.....)))).)))))..)).. (-33.56 = -33.52 + -0.04)

| Location | 1,353,089 – 1,353,209 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -33.48 |
| Consensus MFE | -31.12 |
| Energy contribution | -30.88 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.665280 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1353089 120 - 20766785 AGCGUUCGCUCAUUUUAAUUACAGAACUAUGUGGCUUUCUGAGCUCGCCGAUUCAGAAUUUCUGAUUUCGCGCUGCAGGCCGCAAGUUUGCCAUGGUAUUUAAUUAACGCCGCCGCUCUC ((((...((.....((((.(((.(((((.((((((((.....((.(((.((.((((.....)))).)).)))..)))))))))))))))......))).)))).....))...))))... ( -32.90) >DroSec_CAF1 71219 119 - 1 AGCGUUCGCUCAUUUUAAUUACAGAACUAUGUGGCUUUCUGAGCUCGCCGAUUCAGAAUUUCUGAU-UCGCGCUGCAGGCCGCAAGUUUGCCAUGGUAUUUAAUUAACGCCGCUGCUCUC ((((...((.....((((.(((.(((((.((((((((.....((..(((((.((((.....)))).-))).)).)))))))))))))))......))).)))).....))))))...... ( -33.80) >DroSim_CAF1 49387 119 - 1 AGCGUUCGCUCAUUUUAAUUACAGAACUAUGUGGCUUUCUGAGCUCGCCGAUUCAGAAUUUCUGAU-UCGCGCUGCAGGCCGCAAGUUUGCCAUGGUAUUUAAUUAACGCCGCUGCUCUC ((((...((.....((((.(((.(((((.((((((((.....((..(((((.((((.....)))).-))).)).)))))))))))))))......))).)))).....))))))...... ( -33.80) >DroEre_CAF1 51842 120 - 1 AGCGUUCGCUCAUUUUAAUUACAGAACUAUGUGGCUUUCUGAGCCCGCCGAUUCAGAAUUUCUGAUUUCGCGCUGCAGGCCGCAAGUUUGCCAUGGUAUUUAAUUAACGCCGCCGCGCUC (((((..((..............(((((.((((((...((((((..((.((.((((.....)))).)).))))).)))))))))))))).....((..((.....))..)))).))))). ( -34.60) >DroYak_CAF1 39102 120 - 1 AGCGUUCGCUCAUUUUAAUUACAGAACUAUGUGGCUUUCUGAGCUCGCCGAUUCAGAAUUUCUGAUUUCGCGCUGCAGGCCGCAAGUUUGCCAUGGUAUUUAAUUAACGCCGCUGCUCUC ((((...((.....((((.(((.(((((.((((((((.....((.(((.((.((((.....)))).)).)))..)))))))))))))))......))).)))).....))))))...... ( -32.30) >consensus AGCGUUCGCUCAUUUUAAUUACAGAACUAUGUGGCUUUCUGAGCUCGCCGAUUCAGAAUUUCUGAUUUCGCGCUGCAGGCCGCAAGUUUGCCAUGGUAUUUAAUUAACGCCGCUGCUCUC .(((...((.....((((.(((.(((((.((((((...((((((.((.....((((.....))))...)).))).))))))))))))))......))).)))).....))...))).... (-31.12 = -30.88 + -0.24)

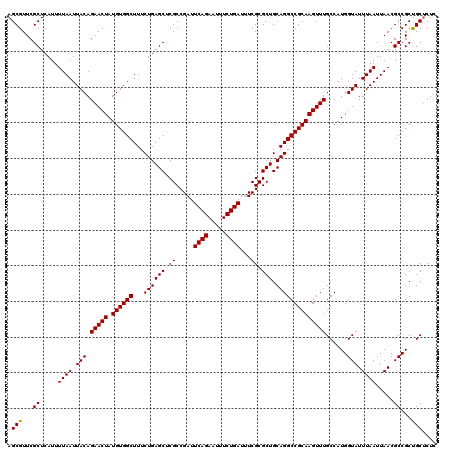
| Location | 1,353,129 – 1,353,249 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.50 |
| Mean single sequence MFE | -30.65 |
| Consensus MFE | -28.93 |
| Energy contribution | -28.93 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.867850 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1353129 120 - 20766785 CACAUAAAUUAAAAUUGAAAUUCGGCCGCAGAAUUGCCAAAGCGUUCGCUCAUUUUAAUUACAGAACUAUGUGGCUUUCUGAGCUCGCCGAUUCAGAAUUUCUGAUUUCGCGCUGCAGGC ........................(((.(((((..((((....((((................))))....)))).))))).((.(((.((.((((.....)))).)).)))..)).))) ( -30.09) >DroSec_CAF1 71259 119 - 1 CACAUAAAUUAAAAUUGAAACUCGGCCGCAGAAUUGCCAAAGCGUUCGCUCAUUUUAAUUACAGAACUAUGUGGCUUUCUGAGCUCGCCGAUUCAGAAUUUCUGAU-UCGCGCUGCAGGC ........................(((.(((((..((((....((((................))))....)))).))))).((..(((((.((((.....)))).-))).)).)).))) ( -31.59) >DroSim_CAF1 49427 119 - 1 CACAUAAAUUAAAAUUGAAACUCGGCCGCAGAAUUGCCAAAGCGUUCGCUCAUUUUAAUUACAGAACUAUGUGGCUUUCUGAGCUCGCCGAUUCAGAAUUUCUGAU-UCGCGCUGCAGGC ........................(((.(((((..((((....((((................))))....)))).))))).((..(((((.((((.....)))).-))).)).)).))) ( -31.59) >DroEre_CAF1 51882 120 - 1 CACAUAAAUUAAAAUUGAAACUCGGCCGCAGAAUUGCCAAAGCGUUCGCUCAUUUUAAUUACAGAACUAUGUGGCUUUCUGAGCCCGCCGAUUCAGAAUUUCUGAUUUCGCGCUGCAGGC ........................(((.(((((..((((....((((................))))....)))).))))).((..(((((.((((.....))))..))).)).)).))) ( -29.59) >DroYak_CAF1 39142 120 - 1 CACAUAAAUUAAAAUUGAAACUCGGCCACAGAAUUGCCAAAGCGUUCGCUCAUUUUAAUUACAGAACUAUGUGGCUUUCUGAGCUCGCCGAUUCAGAAUUUCUGAUUUCGCGCUGCAGGC ........................(((.(((((..((((....((((................))))....)))).))))).((.(((.((.((((.....)))).)).)))..)).))) ( -30.39) >consensus CACAUAAAUUAAAAUUGAAACUCGGCCGCAGAAUUGCCAAAGCGUUCGCUCAUUUUAAUUACAGAACUAUGUGGCUUUCUGAGCUCGCCGAUUCAGAAUUUCUGAUUUCGCGCUGCAGGC ........................(((.(((((..((((....((((................))))....)))).))))).((..(((((.((((.....))))..))).)).)).))) (-28.93 = -28.93 + -0.00)

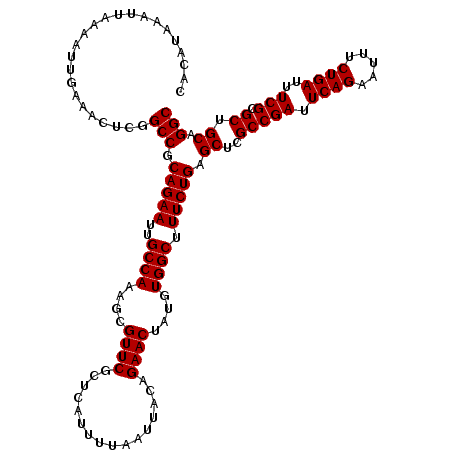
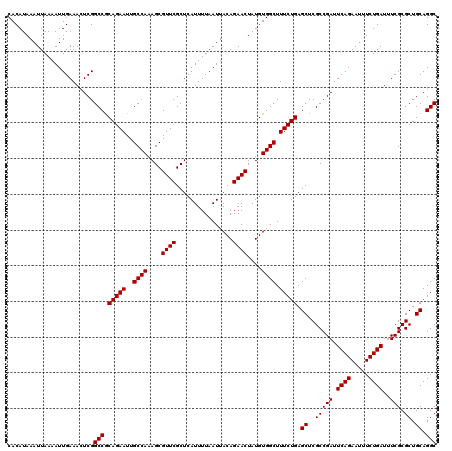
| Location | 1,353,169 – 1,353,289 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.38 |
| Mean single sequence MFE | -26.09 |
| Consensus MFE | -24.11 |
| Energy contribution | -24.19 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.775400 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1353169 120 - 20766785 CAAACAGCGCUAAUGAAUACAAAUUUCGCGAAGCGCUGUGCACAUAAAUUAAAAUUGAAAUUCGGCCGCAGAAUUGCCAAAGCGUUCGCUCAUUUUAAUUACAGAACUAUGUGGCUUUCU ...((((((((..((((.......))))...)))))))).((((((......(((((((((.((..(((............)))..))...))))))))).......))))))....... ( -29.22) >DroSec_CAF1 71298 116 - 1 ----CAGCGCUAAUGAAUACAAAUUUCGCGAAGCGCUGUGCACAUAAAUUAAAAUUGAAACUCGGCCGCAGAAUUGCCAAAGCGUUCGCUCAUUUUAAUUACAGAACUAUGUGGCUUUCU ----(((((((..((((.......))))...)))))))..(((((((((((((((.((....((..(((............)))..)).))))))))))).......))))))....... ( -26.21) >DroSim_CAF1 49466 116 - 1 ----CAGCGCUAAUGAAUACAAAUUUCGCGAAGCGCUGUGCACAUAAAUUAAAAUUGAAACUCGGCCGCAGAAUUGCCAAAGCGUUCGCUCAUUUUAAUUACAGAACUAUGUGGCUUUCU ----(((((((..((((.......))))...)))))))..(((((((((((((((.((....((..(((............)))..)).))))))))))).......))))))....... ( -26.21) >DroEre_CAF1 51922 120 - 1 AAAGCAGCUCUAAUGAAUACAAAUUUCACGAAGCGCUGUGCACAUAAAUUAAAAUUGAAACUCGGCCGCAGAAUUGCCAAAGCGUUCGCUCAUUUUAAUUACAGAACUAUGUGGCUUUCU ...(((((.((..((((.......))))...)).))))).(((((((((((((((.((....((..(((............)))..)).))))))))))).......))))))....... ( -23.51) >DroYak_CAF1 39182 116 - 1 A---CUGCGCUAAUGAAUACAAAUUUCGAG-AGCGCUGUGCACAUAAAUUAAAAUUGAAACUCGGCCACAGAAUUGCCAAAGCGUUCGCUCAUUUUAAUUACAGAACUAUGUGGCUUUCU (---(.(((((..((((.......))))..-))))).))........................(((((((.....((....))((((................))))..))))))).... ( -25.29) >consensus ____CAGCGCUAAUGAAUACAAAUUUCGCGAAGCGCUGUGCACAUAAAUUAAAAUUGAAACUCGGCCGCAGAAUUGCCAAAGCGUUCGCUCAUUUUAAUUACAGAACUAUGUGGCUUUCU ....(((((((..((((.......))))...))))))).........................(((((((.....((....))((((................))))..))))))).... (-24.11 = -24.19 + 0.08)

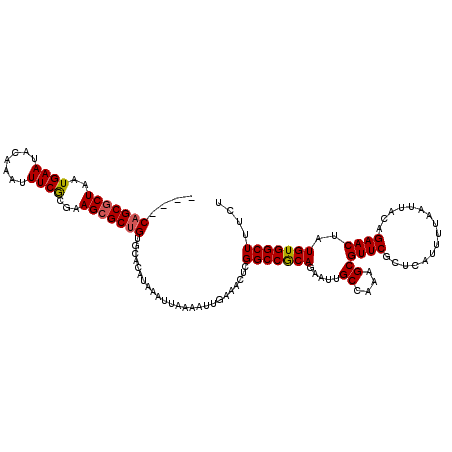
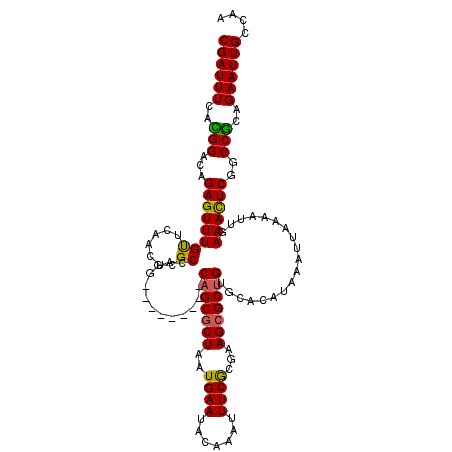
| Location | 1,353,209 – 1,353,329 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.84 |
| Mean single sequence MFE | -30.82 |
| Consensus MFE | -23.16 |
| Energy contribution | -22.68 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.812160 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1353209 120 - 20766785 CGAUUUCACGGACAGAGUUUGUUCAACUCGCCAGGCCAGGCAAACAGCGCUAAUGAAUACAAAUUUCGCGAAGCGCUGUGCACAUAAAUUAAAAUUGAAAUUCGGCCGCAGAAUUGCCAA ((((((..(((...(((((.....))))).)).((((..((..((((((((..((((.......))))...)))))))))).((...........))......)))))..)))))).... ( -32.50) >DroSec_CAF1 71338 110 - 1 CGAUUUCGCGGACAGAGUUUGUUCAACUCGCCAG----------CAGCGCUAAUGAAUACAAAUUUCGCGAAGCGCUGUGCACAUAAAUUAAAAUUGAAACUCGGCCGCAGAAUUGCCAA (((((..((((...((((((.........((..(----------(((((((..((((.......))))...)))))))))).((...........))))))))..))))..))))).... ( -33.50) >DroSim_CAF1 49506 110 - 1 CGAUUUCGCGGACAGAGUUUGUUCAACUCGCCAG----------CAGCGCUAAUGAAUACAAAUUUCGCGAAGCGCUGUGCACAUAAAUUAAAAUUGAAACUCGGCCGCAGAAUUGCCAA (((((..((((...((((((.........((..(----------(((((((..((((.......))))...)))))))))).((...........))))))))..))))..))))).... ( -33.50) >DroEre_CAF1 51962 118 - 1 CGAUUUCAUGGACAGAGUUUGCUCCACUGGCCGGGA--AAAAAGCAGCUCUAAUGAAUACAAAUUUCACGAAGCGCUGUGCACAUAAAUUAAAAUUGAAACUCGGCCGCAGAAUUGCCAA ........(((.(((..(((((......(((((((.--.....(((((.((..((((.......))))...)).)))))...((...........))...)))))))))))).)))))). ( -29.00) >DroYak_CAF1 39222 114 - 1 CGAUUUCAUGGACAGAGUUUGUUCAACUCGCUGCGA--AAA---CUGCGCUAAUGAAUACAAAUUUCGAG-AGCGCUGUGCACAUAAAUUAAAAUUGAAACUCGGCCACAGAAUUGCCAA ((((((..(((...((((((...........((((.--...---..(((((..((((.......))))..-)))))..))))((...........))))))))..)))..)))))).... ( -25.60) >consensus CGAUUUCACGGACAGAGUUUGUUCAACUCGCCAGG_________CAGCGCUAAUGAAUACAAAUUUCGCGAAGCGCUGUGCACAUAAAUUAAAAUUGAAACUCGGCCGCAGAAUUGCCAA ((((((..(((...((((((((.......)).............(((((((..((((.......))))...)))))))...................))))))..)))..)))))).... (-23.16 = -22.68 + -0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:27:35 2006