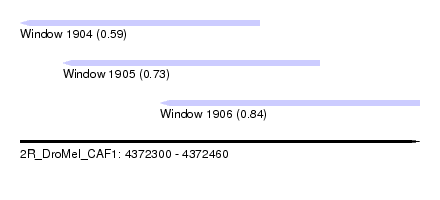
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,372,300 – 4,372,460 |
| Length | 160 |
| Max. P | 0.840588 |

| Location | 4,372,300 – 4,372,396 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 94.79 |
| Mean single sequence MFE | -37.45 |
| Consensus MFE | -33.13 |
| Energy contribution | -33.17 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.587512 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
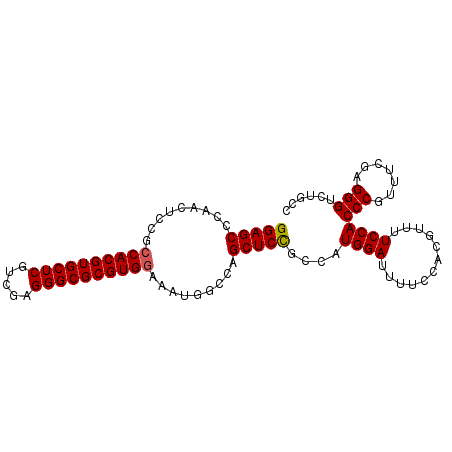
>2R_DroMel_CAF1 4372300 96 - 20766785 GGAGCCCAACUCCGCCACGUGCUCGUUGAGGGCGCGUGGAAAUAGCCAGCUCCGCCAUGGAUUUUCCACGUUUUCCACCCGUUUCGAGGGUCUGCC (((((.........((((((((((.....)))))))))).........)))))((..(((.....)))........((((.......))))..)). ( -36.27) >DroSec_CAF1 42962 96 - 1 GGAGCCCAACUCCGCCACGUGCUCGUCGAGGGCGCGUGGAAAUGGGCAGCUCCGCCAUGGAUUUUCCACGUUUUCCACCCGUUUCGAGGGUCUGCC ((((((((......((((((((((.....))))))))))...)))))..........(((.....))).....)))((((.......))))..... ( -38.50) >DroSim_CAF1 43195 96 - 1 GGAGCCCAACUCCGCCACGUGCUCGUCGAGGGCGCGUGGAAAUGGCCAGCUCCGCCAUGGAUUUUCCACGUUUUCCACCCGUUUCGAGGGUCUGCC ((((((((......((((((((((.....))))))))))...)))...)))))((..(((.....)))........((((.......))))..)). ( -37.00) >DroEre_CAF1 39476 95 - 1 GGAGCCCUACUCCGCCACGUGCUCGUCGAGGGCGCGUGGAAGUGGCCAGCUCUGGCAUGGAUUUUUCACGUUUUCCACCCGUUUCGAGGGGCU-CC (((((((((((...((((((((((.....)))))))))).))).((((....)))).((((............))))..........))))))-)) ( -43.90) >DroYak_CAF1 41386 96 - 1 GGAGCCCAACUCCGCCACGUGCUCCUUGAGGGCGCGUGAAAAUGGCCAGCUCCGCCAUGGAUUUUCCACGUUUUCCACCCGUUUCGAGGGUCUGCC ((((.....))))..(((((((((.....)))))))))...(((((.......)))))(((............)))((((.......))))..... ( -31.60) >consensus GGAGCCCAACUCCGCCACGUGCUCGUCGAGGGCGCGUGGAAAUGGCCAGCUCCGCCAUGGAUUUUCCACGUUUUCCACCCGUUUCGAGGGUCUGCC (((((.........((((((((((.....)))))))))).........)))))....((((............))))(((.......)))...... (-33.13 = -33.17 + 0.04)

| Location | 4,372,317 – 4,372,420 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 92.91 |
| Mean single sequence MFE | -34.39 |
| Consensus MFE | -29.50 |
| Energy contribution | -30.14 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.734850 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
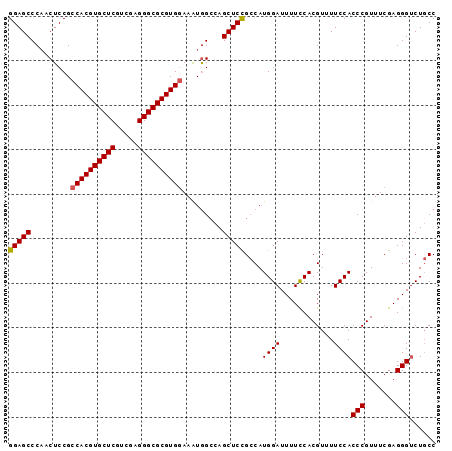
>2R_DroMel_CAF1 4372317 103 - 20766785 GAACACCACCACAAUUUUCCGCCUGGAGCCCAACUCCGCCACGUGCUCGUUGAGGGCGCGUGGAAAUAGCCAGCUCCGCCAUGGAUUUUCCACGUUUUCCACC ((((....................(((((.........((((((((((.....)))))))))).........)))))....(((.....))).))))...... ( -31.97) >DroSec_CAF1 42979 103 - 1 GCACACCACCGCCAUUUUCCGCCUGGAGCCCAACUCCGCCACGUGCUCGUCGAGGGCGCGUGGAAAUGGGCAGCUCCGCCAUGGAUUUUCCACGUUUUCCACC ((...(((..((........)).))).(((((......((((((((((.....))))))))))...))))).)).......((((............)))).. ( -34.80) >DroSim_CAF1 43212 103 - 1 GCACACCACCGCCAUUUUCCGCCUGGAGCCCAACUCCGCCACGUGCUCGUCGAGGGCGCGUGGAAAUGGCCAGCUCCGCCAUGGAUUUUCCACGUUUUCCACC ..........(((((((.......((((.....)))).((((((((((.....)))))))))))))))))...........((((............)))).. ( -34.50) >DroEre_CAF1 39492 103 - 1 GCACAACACCGCCAUUUUCGCCCUGGAGCCCUACUCCGCCACGUGCUCGUCGAGGGCGCGUGGAAGUGGCCAGCUCUGGCAUGGAUUUUUCACGUUUUCCACC ((........)).......(((..(((((.(((((...((((((((((.....)))))))))).)))))...)))))))).((((............)))).. ( -37.90) >DroYak_CAF1 41403 102 - 1 GCACA-CGCCGCCAUUUUCCGCCUGGAGCCCAACUCCGCCACGUGCUCCUUGAGGGCGCGUGAAAAUGGCCAGCUCCGCCAUGGAUUUUCCACGUUUUCCACC .....-.((.(((((((((.....((((.....))))...((((((((.....)))))))))))))))))..)).......((((............)))).. ( -32.80) >consensus GCACACCACCGCCAUUUUCCGCCUGGAGCCCAACUCCGCCACGUGCUCGUCGAGGGCGCGUGGAAAUGGCCAGCUCCGCCAUGGAUUUUCCACGUUUUCCACC ..........(((((((.......((((.....)))).((((((((((.....)))))))))))))))))...........((((............)))).. (-29.50 = -30.14 + 0.64)

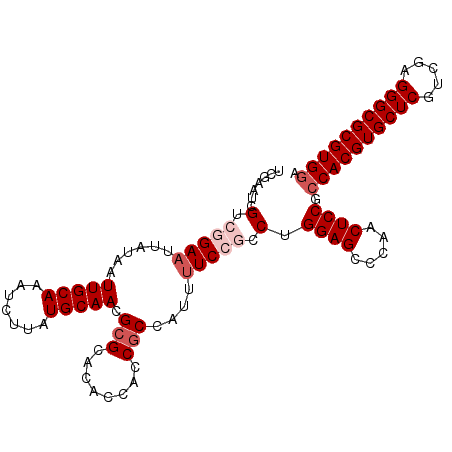
| Location | 4,372,356 – 4,372,460 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 94.13 |
| Mean single sequence MFE | -32.40 |
| Consensus MFE | -28.42 |
| Energy contribution | -29.82 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.840588 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
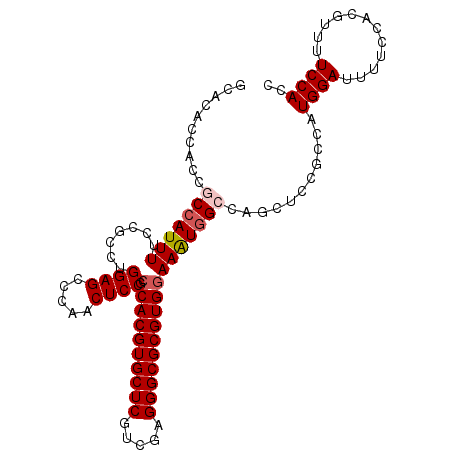
>2R_DroMel_CAF1 4372356 104 - 20766785 UCGAAUCGUCGGAAUUAUAAUUGCAAAUCUUAUGCAACGCGAACACCACCACAAUUUUCCGCCUGGAGCCCAACUCCGCCACGUGCUCGUUGAGGGCGCGUGGA .......(.(((((......(((((.......))))).(.(........).)....))))).).((((.....)))).((((((((((.....)))))))))). ( -31.10) >DroSec_CAF1 43018 104 - 1 UCGAAUCGUCGGAAUUAUAAUUGCAAAUCUUAUGCAACGCGCACACCACCGCCAUUUUCCGCCUGGAGCCCAACUCCGCCACGUGCUCGUCGAGGGCGCGUGGA .......(.(((((......(((((.......))))).(((........)))....))))).).((((.....)))).((((((((((.....)))))))))). ( -34.80) >DroSim_CAF1 43251 104 - 1 UCGAAUCGUCGGAAUUAUAAUUGCAAAUCUUAUGCAACGCGCACACCACCGCCAUUUUCCGCCUGGAGCCCAACUCCGCCACGUGCUCGUCGAGGGCGCGUGGA .......(.(((((......(((((.......))))).(((........)))....))))).).((((.....)))).((((((((((.....)))))))))). ( -34.80) >DroEre_CAF1 39531 104 - 1 UCGAAUCGUCGGAAUUAUAAUUGCAAAUCUUAUGCAACGCGCACAACACCGCCAUUUUCGCCCUGGAGCCCUACUCCGCCACGUGCUCGUCGAGGGCGCGUGGA .......(.(((((......(((((.......))))).(((........)))...))))))...((((.....)))).((((((((((.....)))))))))). ( -31.50) >DroYak_CAF1 41442 103 - 1 UCGAAUCGUAAGAUUUAUAAUUGCAAAUCUUAUGCAACGCGCACA-CGCCGCCAUUUUCCGCCUGGAGCCCAACUCCGCCACGUGCUCCUUGAGGGCGCGUGAA ..(((((....)))))....(((((.......))))).(((....-)))...............((((.....))))..(((((((((.....))))))))).. ( -29.80) >consensus UCGAAUCGUCGGAAUUAUAAUUGCAAAUCUUAUGCAACGCGCACACCACCGCCAUUUUCCGCCUGGAGCCCAACUCCGCCACGUGCUCGUCGAGGGCGCGUGGA .......(.(((((......(((((.......))))).(((........)))....))))).).((((.....)))).((((((((((.....)))))))))). (-28.42 = -29.82 + 1.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:54 2006