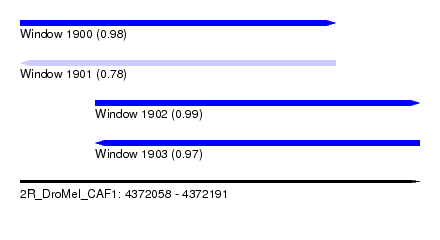
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,372,058 – 4,372,191 |
| Length | 133 |
| Max. P | 0.990721 |

| Location | 4,372,058 – 4,372,163 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 89.94 |
| Mean single sequence MFE | -41.44 |
| Consensus MFE | -35.24 |
| Energy contribution | -35.72 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.982421 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4372058 105 + 20766785 UUCGGCAAGAGCCGACCCACUGUGCCGACUGCUGCGUACUGCGUGCCGCAGUGCCUUCUGCGCUCGCCGAGCCGCCAUUCUCGCCGCUCGGCACCCAUGCCACAU .(((((....))))).....((((.(((.(((.(.((((((((...)))))))))....))).))(((((((.((.......)).)))))))......).)))). ( -43.00) >DroSec_CAF1 42720 105 + 1 UCCUGCAAAAACCGACCCACUGUGCCGGCUGCUGCGUACUGCGUGCCGCAGUGCCUUCUGCGCUCGCCGAGCGUCCAUUCUCGCCGCUCGGCACCCAUGCCACAU ....(((..............((((.(((.(((((((((...))).)))))))))....))))..((((((((.(.......).)))))))).....)))..... ( -36.50) >DroSim_CAF1 42953 105 + 1 UCCUGCGAGAACCGACCCACUGUGCCGGCUGCUGCGUACUGCGUGCCGCAGUGCCUUCUGCGCUCGCCGAGCGUCCAUUCUCGCCGCUCGGCACCCAUGCCACAU ....(((((((..(((.......((((((.((.((((((((((...)))))))).....))))..)))).)))))..))))))).....((((....)))).... ( -39.41) >DroEre_CAF1 39240 99 + 1 ------AAUACCCGACCCACUGUGCCGGCUGCUGCGUACUGCGUGCCGCAGUGCCUUCUGCGCUCGCCGAGCGGCCAUUCUCGCCGCUCGGCACCCAUGCCACAU ------...............((((((((.((.((((((((((...)))))))).....))))..)))(((((((.......))))))))))))........... ( -41.60) >DroYak_CAF1 41150 99 + 1 ------GAUUUCUGGCCCACUGUGCGGGCUGCUGCGUAGUGCUUGCCGCAGUGCCUUUUGCGCUCGCCGAGCGGCCAUUCUCGCCGCUCGGCACCCAUGCCACAU ------......((((.....((((((((.((((((..((....))))))))))))...))))..((((((((((.......))))))))))......))))... ( -46.70) >consensus U_C_GCAAGAACCGACCCACUGUGCCGGCUGCUGCGUACUGCGUGCCGCAGUGCCUUCUGCGCUCGCCGAGCGGCCAUUCUCGCCGCUCGGCACCCAUGCCACAU .....................((((.(((.(((((((((...))).)))))))))....))))..((((((((((.......))))))))))............. (-35.24 = -35.72 + 0.48)

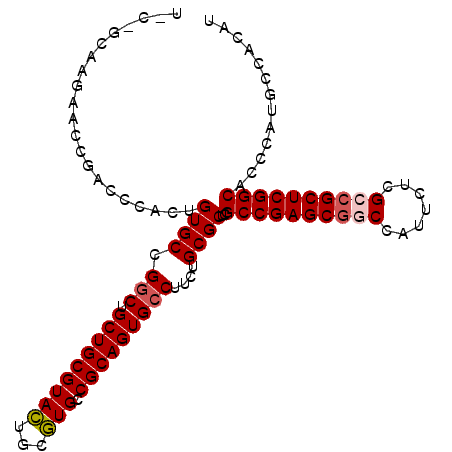
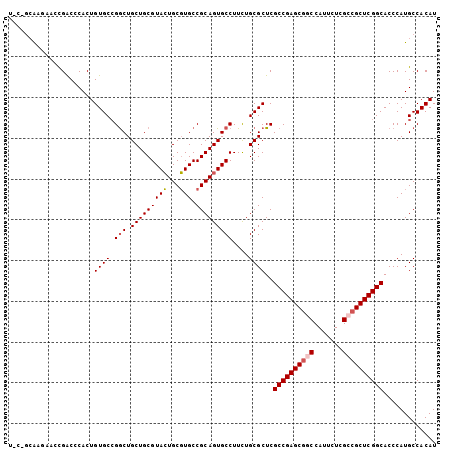
| Location | 4,372,058 – 4,372,163 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 89.94 |
| Mean single sequence MFE | -45.70 |
| Consensus MFE | -38.24 |
| Energy contribution | -38.76 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.777698 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4372058 105 - 20766785 AUGUGGCAUGGGUGCCGAGCGGCGAGAAUGGCGGCUCGGCGAGCGCAGAAGGCACUGCGGCACGCAGUACGCAGCAGUCGGCACAGUGGGUCGGCUCUUGCCGAA .((((((.....((((((((.((.......)).))))))))..(((((......))))))).))))....((((.(((((((.......))))))).)))).... ( -46.70) >DroSec_CAF1 42720 105 - 1 AUGUGGCAUGGGUGCCGAGCGGCGAGAAUGGACGCUCGGCGAGCGCAGAAGGCACUGCGGCACGCAGUACGCAGCAGCCGGCACAGUGGGUCGGUUUUUGCAGGA .((((((.....(((((((((...........)))))))))..(((((......))))))).))))....((((.(((((((.......))))))).)))).... ( -45.60) >DroSim_CAF1 42953 105 - 1 AUGUGGCAUGGGUGCCGAGCGGCGAGAAUGGACGCUCGGCGAGCGCAGAAGGCACUGCGGCACGCAGUACGCAGCAGCCGGCACAGUGGGUCGGUUCUCGCAGGA ...(((((....)))))....((((((((.(((((.((((..((((.....))((((((...)))))).....)).)))))).(....)))).)))))))).... ( -44.60) >DroEre_CAF1 39240 99 - 1 AUGUGGCAUGGGUGCCGAGCGGCGAGAAUGGCCGCUCGGCGAGCGCAGAAGGCACUGCGGCACGCAGUACGCAGCAGCCGGCACAGUGGGUCGGGUAUU------ ((.((((....((((((((((((.......)))))))(((..((((.....))((((((...)))))).....)).)))))))).....)))).))...------ ( -46.40) >DroYak_CAF1 41150 99 - 1 AUGUGGCAUGGGUGCCGAGCGGCGAGAAUGGCCGCUCGGCGAGCGCAAAAGGCACUGCGGCAAGCACUACGCAGCAGCCCGCACAGUGGGCCAGAAAUC------ .(.((((.((..(((((((((((.......))))))))))).(((.....(((.(((((..........)))))..)))))).))....)))).)....------ ( -45.20) >consensus AUGUGGCAUGGGUGCCGAGCGGCGAGAAUGGCCGCUCGGCGAGCGCAGAAGGCACUGCGGCACGCAGUACGCAGCAGCCGGCACAGUGGGUCGGUUAUUGC_G_A ...((((....((((((((((((.......)))))))(((..((((.....))(((((.....))))).....)).)))))))).....))))............ (-38.24 = -38.76 + 0.52)

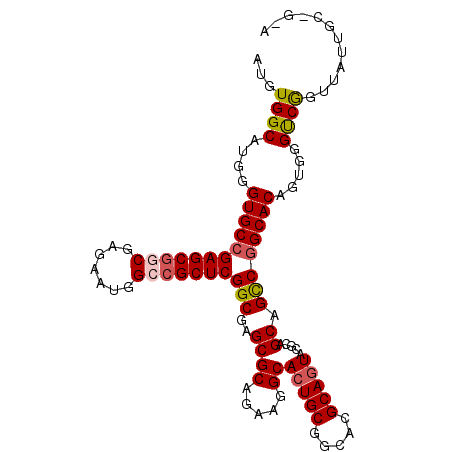
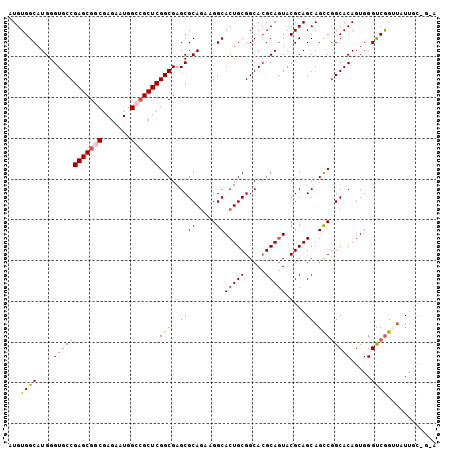
| Location | 4,372,083 – 4,372,191 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 96.02 |
| Mean single sequence MFE | -42.08 |
| Consensus MFE | -38.58 |
| Energy contribution | -39.42 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.990721 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4372083 108 + 20766785 CGACUGCUGCGUACUGCGUGCCGCAGUGCCUUCUGCGCUCGCCGAGCCGCCAUUCUCGCCGCUCGGCACCCAUGCCACAUCGCAUCGCCUAUCAUUCUGCACUCGUGG (((.(((...((((((((...)))))))).....(((...(((((((.((.......)).)))))))....((((......)))))))..........))).)))... ( -38.40) >DroSec_CAF1 42745 108 + 1 CGGCUGCUGCGUACUGCGUGCCGCAGUGCCUUCUGCGCUCGCCGAGCGUCCAUUCUCGCCGCUCGGCACCCAUGCCACAUCGCAUCGCCUAUCAUUCUGCACUCGUGG .(((.(.((((....(((((.(((((......)))))...((((((((.(.......).))))))))...))))).....)))).)))).......(..(....)..) ( -39.00) >DroSim_CAF1 42978 108 + 1 CGGCUGCUGCGUACUGCGUGCCGCAGUGCCUUCUGCGCUCGCCGAGCGUCCAUUCUCGCCGCUCGGCACCCAUGCCACAUCGCAUCGCCUAUCAUUCUGCACUCGUGG .(((.(.((((....(((((.(((((......)))))...((((((((.(.......).))))))))...))))).....)))).)))).......(..(....)..) ( -39.00) >DroEre_CAF1 39259 108 + 1 CGGCUGCUGCGUACUGCGUGCCGCAGUGCCUUCUGCGCUCGCCGAGCGGCCAUUCUCGCCGCUCGGCACCCAUGCCACAUCGCAUUGCCCAUCAUUCUGCACUCGUGG .(((...((((....(((((.(((((......)))))...((((((((((.......))))))))))...))))).....))))..))).......(..(....)..) ( -45.90) >DroYak_CAF1 41169 108 + 1 GGGCUGCUGCGUAGUGCUUGCCGCAGUGCCUUUUGCGCUCGCCGAGCGGCCAUUCUCGCCGCUCGGCACCCAUGCCACAUCGCAUGGCCCAUCAUUCUGCACUCGUGG ......(..((.(((((.....(.(((((.....))))))((((((((((.......))))))))))..((((((......))))))...........)))))))..) ( -48.10) >consensus CGGCUGCUGCGUACUGCGUGCCGCAGUGCCUUCUGCGCUCGCCGAGCGGCCAUUCUCGCCGCUCGGCACCCAUGCCACAUCGCAUCGCCUAUCAUUCUGCACUCGUGG .(((...((((....(((((.(((((......)))))...((((((((((.......))))))))))...))))).....))))..))).......(..(....)..) (-38.58 = -39.42 + 0.84)

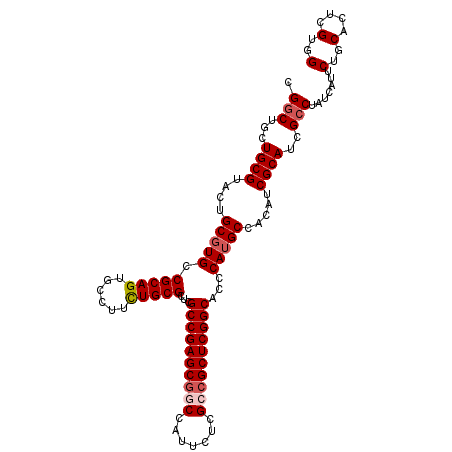
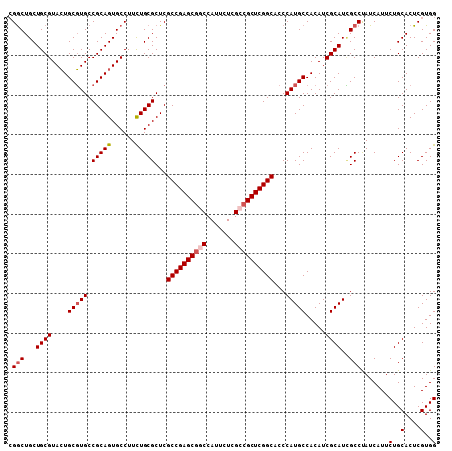
| Location | 4,372,083 – 4,372,191 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 96.02 |
| Mean single sequence MFE | -46.56 |
| Consensus MFE | -42.60 |
| Energy contribution | -43.44 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.969976 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4372083 108 - 20766785 CCACGAGUGCAGAAUGAUAGGCGAUGCGAUGUGGCAUGGGUGCCGAGCGGCGAGAAUGGCGGCUCGGCGAGCGCAGAAGGCACUGCGGCACGCAGUACGCAGCAGUCG ...(((.(((....((.....)).((((.((((((.....((((((((.((.......)).))))))))..(((((......))))))).))))...))))))).))) ( -45.00) >DroSec_CAF1 42745 108 - 1 CCACGAGUGCAGAAUGAUAGGCGAUGCGAUGUGGCAUGGGUGCCGAGCGGCGAGAAUGGACGCUCGGCGAGCGCAGAAGGCACUGCGGCACGCAGUACGCAGCAGCCG ...................(((..((((.((((((.....(((((((((...........)))))))))..(((((......))))))).))))...))))...))). ( -43.70) >DroSim_CAF1 42978 108 - 1 CCACGAGUGCAGAAUGAUAGGCGAUGCGAUGUGGCAUGGGUGCCGAGCGGCGAGAAUGGACGCUCGGCGAGCGCAGAAGGCACUGCGGCACGCAGUACGCAGCAGCCG ...................(((..((((.((((((.....(((((((((...........)))))))))..(((((......))))))).))))...))))...))). ( -43.70) >DroEre_CAF1 39259 108 - 1 CCACGAGUGCAGAAUGAUGGGCAAUGCGAUGUGGCAUGGGUGCCGAGCGGCGAGAAUGGCCGCUCGGCGAGCGCAGAAGGCACUGCGGCACGCAGUACGCAGCAGCCG ...................(((..((((.((((((.....(((((((((((.......)))))))))))..(((((......))))))).))))...))))...))). ( -49.00) >DroYak_CAF1 41169 108 - 1 CCACGAGUGCAGAAUGAUGGGCCAUGCGAUGUGGCAUGGGUGCCGAGCGGCGAGAAUGGCCGCUCGGCGAGCGCAAAAGGCACUGCGGCAAGCACUACGCAGCAGCCC ......((((...........((((((......)))))).(((((((((((.......))))))))))).))))....(((.(((((..........)))))..))). ( -51.40) >consensus CCACGAGUGCAGAAUGAUAGGCGAUGCGAUGUGGCAUGGGUGCCGAGCGGCGAGAAUGGCCGCUCGGCGAGCGCAGAAGGCACUGCGGCACGCAGUACGCAGCAGCCG ...................(((..((((.((((((.....(((((((((((.......)))))))))))..(((((......))))))).))))...))))...))). (-42.60 = -43.44 + 0.84)

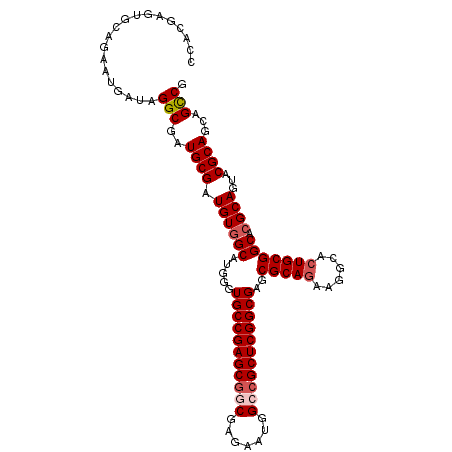
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:51 2006