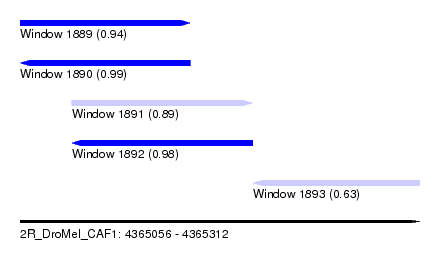
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,365,056 – 4,365,312 |
| Length | 256 |
| Max. P | 0.985894 |

| Location | 4,365,056 – 4,365,165 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 86.03 |
| Mean single sequence MFE | -41.10 |
| Consensus MFE | -30.85 |
| Energy contribution | -31.23 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938822 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
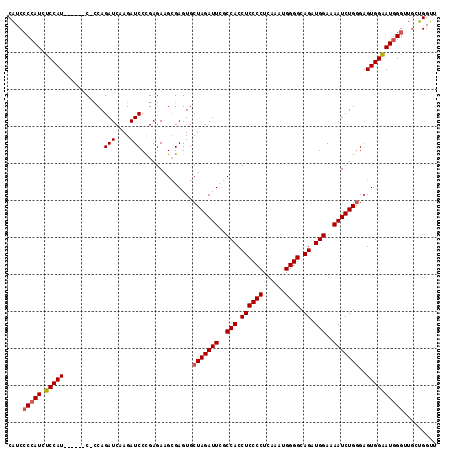
>2R_DroMel_CAF1 4365056 109 + 20766785 CAUCCCCAUCUCCAU------C-CCAGAUCAAGAUCCCGAGAAGCGAGUGCUAGAUUUGCCACCUCCCCUAAAAUGGGGCAGAUGGAAAAUCUGGGAGUGGAAUGGGUUGCUGGUU ....(((((.(((((------(-((((((.....(((......((((((.....))))))...((((((......)))).))..)))..)))))))).)))))))))......... ( -39.20) >DroSec_CAF1 35604 109 + 1 CAUCCCCAUCUCCAU------C-ACAGAUCUAGAUCCUGAGAAGGGAGUGCUAGAUUCGCCACCUCCCCUCAAAUGGGGCAGAUGGAAAAUCUGGGAGUGGAAUGGGCUGCUGGUU ....(((((.(((((------.-.(((((((((.((((.....))))...)))))....(((.((((((......)))).)).)))....))))...))))))))))......... ( -40.20) >DroSim_CAF1 35811 115 + 1 CAUCCCCAUCUCCAUCUCUAUC-CCAGAUCAAGAUCCCGAGAAGCGAGUGCUAGAUUCGCCACCUCCCCUCAAAUGGGGCAGAUGGAAAAUCUGGGAGUGGAAUGGGUUGCUGGUU ....(((((.(((((.....((-((((((.....(((......((((((.....))))))...((((((......)))).))..)))..))))))))))))))))))......... ( -40.40) >DroYak_CAF1 33649 115 + 1 CUUCACAAUACCCAUCCCCAUCCCCAGAUCCAGAUCCCGAGAUGCGAGUGCUAGAUUCGCCACCUCCCCUCAAAUGGGGCAGAUGGAAAAUCUGAGCGUGGGAUGGGA-ACGGGGC ..........(((...(((((((((.(((....))).............((((((((..(((.((((((......)))).)).)))..))))).)))).)))))))).-..))).. ( -44.60) >consensus CAUCCCCAUCUCCAU______C_CCAGAUCAAGAUCCCGAGAAGCGAGUGCUAGAUUCGCCACCUCCCCUCAAAUGGGGCAGAUGGAAAAUCUGGGAGUGGAAUGGGUUGCUGGUU ....(((((.(((((...........(((....)))..............(((((((..(((.((((((......)))).)).)))..)))))))..))))))))))......... (-30.85 = -31.23 + 0.37)

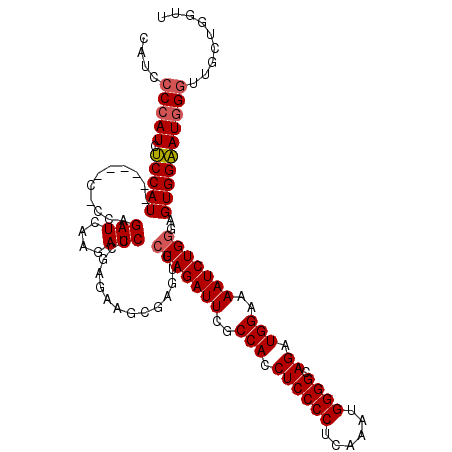
| Location | 4,365,056 – 4,365,165 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 86.03 |
| Mean single sequence MFE | -46.92 |
| Consensus MFE | -33.91 |
| Energy contribution | -33.85 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.41 |
| Structure conservation index | 0.72 |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.985894 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4365056 109 - 20766785 AACCAGCAACCCAUUCCACUCCCAGAUUUUCCAUCUGCCCCAUUUUAGGGGAGGUGGCAAAUCUAGCACUCGCUUCUCGGGAUCUUGAUCUGG-G------AUGGAGAUGGGGAUG .........(((((((((.(((((((((..((((((.((((......))))))))))...........((((.....)))).....)))))))-)------))))).))))).... ( -45.00) >DroSec_CAF1 35604 109 - 1 AACCAGCAGCCCAUUCCACUCCCAGAUUUUCCAUCUGCCCCAUUUGAGGGGAGGUGGCGAAUCUAGCACUCCCUUCUCAGGAUCUAGAUCUGU-G------AUGGAGAUGGGGAUG .........(((((((((.((.(((((((.((((((.((((......)))))))))).)))(((((...(((.......))).))))))))).-)------))))).))))).... ( -40.00) >DroSim_CAF1 35811 115 - 1 AACCAGCAACCCAUUCCACUCCCAGAUUUUCCAUCUGCCCCAUUUGAGGGGAGGUGGCGAAUCUAGCACUCGCUUCUCGGGAUCUUGAUCUGG-GAUAGAGAUGGAGAUGGGGAUG .........(((((((((.((((((((((.((((((.((((......)))))))))).))))))(((....)))....))))((((.(((...-))).)))))))).))))).... ( -44.50) >DroYak_CAF1 33649 115 - 1 GCCCCGU-UCCCAUCCCACGCUCAGAUUUUCCAUCUGCCCCAUUUGAGGGGAGGUGGCGAAUCUAGCACUCGCAUCUCGGGAUCUGGAUCUGGGGAUGGGGAUGGGUAUUGUGAAG ..(((((-(((((((((..(((.((((((.((((((.((((......)))))))))).))))))))).((((.....))))...........)))))))))))))).......... ( -58.20) >consensus AACCAGCAACCCAUUCCACUCCCAGAUUUUCCAUCUGCCCCAUUUGAGGGGAGGUGGCGAAUCUAGCACUCGCUUCUCGGGAUCUUGAUCUGG_G______AUGGAGAUGGGGAUG .........(((((((((.....((((((.((((((.((((......)))))))))).))))))...............((((....))))...........)))).))))).... (-33.91 = -33.85 + -0.06)

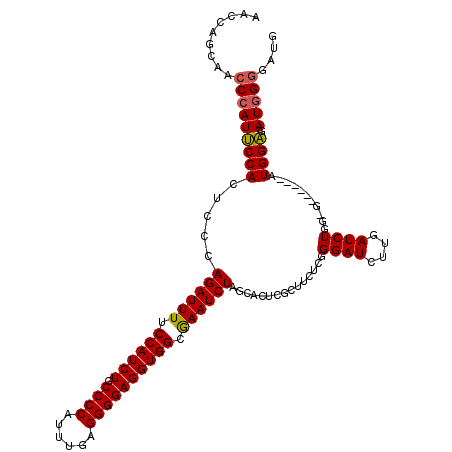
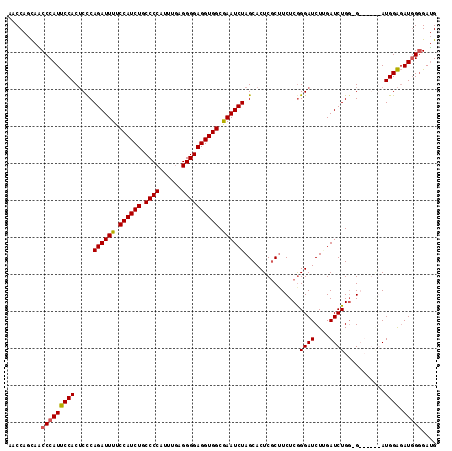
| Location | 4,365,089 – 4,365,205 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 88.99 |
| Mean single sequence MFE | -43.28 |
| Consensus MFE | -31.84 |
| Energy contribution | -34.56 |
| Covariance contribution | 2.72 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886351 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4365089 116 + 20766785 GAAGCGAGUGCUAGAUUUGCCACCUCCCCUAAAAUGGGGCAGAUGGAAAAUCUGGGAGUGGAAUGGGUUGCUGGUUGCUGUGUCGCAACCACAGUGGAGCCUACUUAACUCAAACC ..........((((((((.(((.((((((......)))).)).))).))))))))((((.((.((((((.((((((((......)))))))....).)))))).)).))))..... ( -45.10) >DroSec_CAF1 35637 116 + 1 GAAGGGAGUGCUAGAUUCGCCACCUCCCCUCAAAUGGGGCAGAUGGAAAAUCUGGGAGUGGAAUGGGCUGCUGGUUGCUGUGUCGCAACCACAGUGGAGCCUACUUAACUCAAACC .....((((.(((((((..(((.((((((......)))).)).)))..)))))))((((((..(..((((.(((((((......)))))))))))..)..)))))).))))..... ( -48.90) >DroSim_CAF1 35850 116 + 1 GAAGCGAGUGCUAGAUUCGCCACCUCCCCUCAAAUGGGGCAGAUGGAAAAUCUGGGAGUGGAAUGGGUUGCUGGUUGCUGUGUCGCAACCACAGUGGAGCCUACUUAACUCAAACC ..........(((((((..(((.((((((......)))).)).)))..)))))))((((.((.((((((.((((((((......)))))))....).)))))).)).))))..... ( -45.00) >DroEre_CAF1 32467 109 + 1 GAUGCGAGUGCUAGAUUCGCCACCUCCCCUCAAAUGAGGCAGAUGGAAAAUCUGAGCGUGGGAUGGGA-GCUGGGAGCUGUGUGGCAACCACAGUGGAGCCUACUUAACU------ ...((((((.....))))))...(((((.((..(((...(((((.....)))))..)))..)).))))-).((((.((((((.(....)))))))....)))).......------ ( -39.90) >DroYak_CAF1 33689 109 + 1 GAUGCGAGUGCUAGAUUCGCCACCUCCCCUCAAAUGGGGCAGAUGGAAAAUCUGAGCGUGGGAUGGGA-ACGGGGCGCAGUGUCGCUACCACAGUGGAGCCUACUUAAUU------ (((((..(((((...(((.((((((((((......))))(((((.....))))).....))).)))))-)...))))).)))))(((.((.....)))))..........------ ( -37.50) >consensus GAAGCGAGUGCUAGAUUCGCCACCUCCCCUCAAAUGGGGCAGAUGGAAAAUCUGGGAGUGGAAUGGGAUGCUGGUUGCUGUGUCGCAACCACAGUGGAGCCUACUUAACUCAAACC ..........(((((((..(((.((((((......)))).)).)))..)))))))((((..(.((((((.((((((((......)))))))....).)))))).)..))))..... (-31.84 = -34.56 + 2.72)

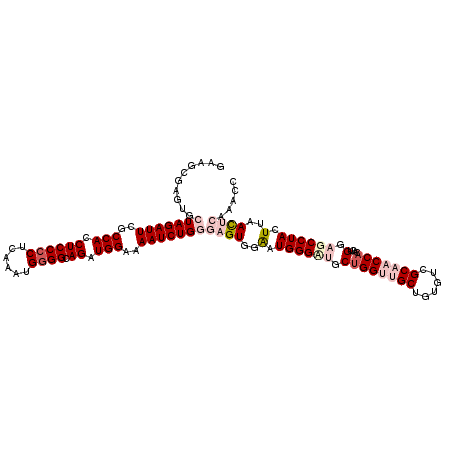
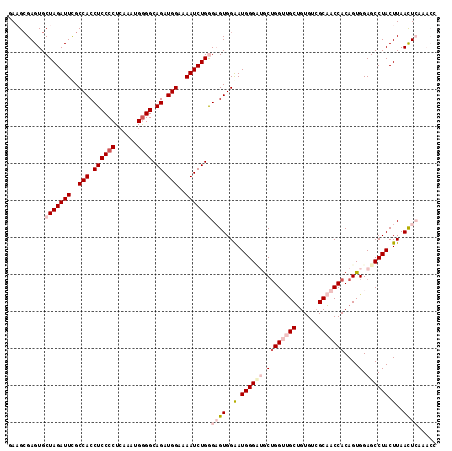
| Location | 4,365,089 – 4,365,205 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 88.99 |
| Mean single sequence MFE | -38.72 |
| Consensus MFE | -31.68 |
| Energy contribution | -32.56 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980496 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4365089 116 - 20766785 GGUUUGAGUUAAGUAGGCUCCACUGUGGUUGCGACACAGCAACCAGCAACCCAUUCCACUCCCAGAUUUUCCAUCUGCCCCAUUUUAGGGGAGGUGGCAAAUCUAGCACUCGCUUC ((..((.(((.(((.(....)))).(((((((......)))))))..))).))..))......((((((.((((((.((((......)))))))))).))))))(((....))).. ( -40.50) >DroSec_CAF1 35637 116 - 1 GGUUUGAGUUAAGUAGGCUCCACUGUGGUUGCGACACAGCAACCAGCAGCCCAUUCCACUCCCAGAUUUUCCAUCUGCCCCAUUUGAGGGGAGGUGGCGAAUCUAGCACUCCCUUC ((...((((..(((.((((.(....(((((((......)))))))).)))).)))..))))))((((((.((((((.((((......)))))))))).))))))............ ( -43.70) >DroSim_CAF1 35850 116 - 1 GGUUUGAGUUAAGUAGGCUCCACUGUGGUUGCGACACAGCAACCAGCAACCCAUUCCACUCCCAGAUUUUCCAUCUGCCCCAUUUGAGGGGAGGUGGCGAAUCUAGCACUCGCUUC ((..((.(((.(((.(....)))).(((((((......)))))))..))).))..))......((((((.((((((.((((......)))))))))).))))))(((....))).. ( -40.90) >DroEre_CAF1 32467 109 - 1 ------AGUUAAGUAGGCUCCACUGUGGUUGCCACACAGCUCCCAGC-UCCCAUCCCACGCUCAGAUUUUCCAUCUGCCUCAUUUGAGGGGAGGUGGCGAAUCUAGCACUCGCAUC ------......(..((((...(((((.......))))).....)))-)..).......(((.((((((.((((((.((((......)))))))))).)))))))))......... ( -33.80) >DroYak_CAF1 33689 109 - 1 ------AAUUAAGUAGGCUCCACUGUGGUAGCGACACUGCGCCCCGU-UCCCAUCCCACGCUCAGAUUUUCCAUCUGCCCCAUUUGAGGGGAGGUGGCGAAUCUAGCACUCGCAUC ------......((..(((((.....)).))).))..((((....((-.........))(((.((((((.((((((.((((......)))))))))).)))))))))...)))).. ( -34.70) >consensus GGUUUGAGUUAAGUAGGCUCCACUGUGGUUGCGACACAGCAACCAGCAACCCAUUCCACUCCCAGAUUUUCCAUCUGCCCCAUUUGAGGGGAGGUGGCGAAUCUAGCACUCGCUUC ............(((((((......(((((((......)))))))..................((((((.((((((.((((......)))))))))).))))))))).)).))... (-31.68 = -32.56 + 0.88)

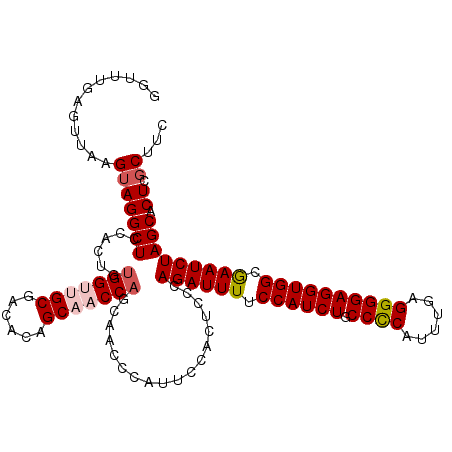
| Location | 4,365,205 – 4,365,312 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 87.17 |
| Mean single sequence MFE | -18.44 |
| Consensus MFE | -14.08 |
| Energy contribution | -13.92 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.628019 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4365205 107 - 20766785 CAAAUAUUUAAAGGAAAGCCAAACCCCAUUUGGCGCAAUAAGAAAUGUAAUAAACCCACAUUUAUGCAAUUAAAAUUUCCCGCACCUAAAGUGUUUAAUUGUCGAGU ...(((.((((.(((((((((((.....))))))(((.....((((((.........)))))).)))........))))).((((.....)))))))).)))..... ( -18.00) >DroSec_CAF1 35753 107 - 1 CAAAUAUUUAAGGGAAAGCCAAACCCCAUUUGGCGCAAUAAGAAAUGUAAUAAACCCACAUUUAUGCAAUUAAAAUUUCCCGCUCCUCAAGUGUUUAAUUAUCGAGU .((((((((..((((((((((((.....))))))(((.....((((((.........)))))).)))........)))))).......))))))))........... ( -21.20) >DroSim_CAF1 35966 107 - 1 CAAAUAUUUAAGGGAAAGCCAAACCCCAUUUGGCGCAAUAAGAAAUGUAAUAAACCCACAUUUAUGCAAUUAAAAUUUCCCGCUCCUCAAGUGUUUAAUUGUCGAGU ...........((((((((((((.....))))))(((.....((((((.........)))))).)))........))))))((((..(((........)))..)))) ( -25.40) >DroEre_CAF1 32576 96 - 1 CAGAUAUUUAAAGGUAAGCCAAACCCCAUUUGGUGCAAUAAGAAAUGUAAUAAACCCACAUUUAUGCAAUUAAACUCUC-CGCUCCGAAAGUGUUUA---------- .((((((((...((...((((((.....))))))(((.....((((((.........)))))).)))............-....))..)))))))).---------- ( -13.90) >DroYak_CAF1 33798 97 - 1 CGAAUAUUUAAAGGGAAACUAAACCCUAUUUGGCGCAAUAAGCAAUGUAAUAAACCCACAUUUAUGCAAUUAAACUGUCCCGCACCUAAAGUGUUUA---------- .((((((((..((((........)))).....((((.....))..((((.((((......)))))))).............)).....)))))))).---------- ( -13.70) >consensus CAAAUAUUUAAAGGAAAGCCAAACCCCAUUUGGCGCAAUAAGAAAUGUAAUAAACCCACAUUUAUGCAAUUAAAAUUUCCCGCUCCUAAAGUGUUUAAUU_UCGAGU .((((((((..(((...((((((.....))))))(((.....((((((.........)))))).))).................))).))))))))........... (-14.08 = -13.92 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:42 2006