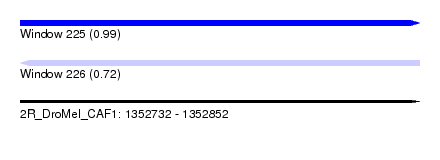
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,352,732 – 1,352,852 |
| Length | 120 |
| Max. P | 0.992408 |

| Location | 1,352,732 – 1,352,852 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
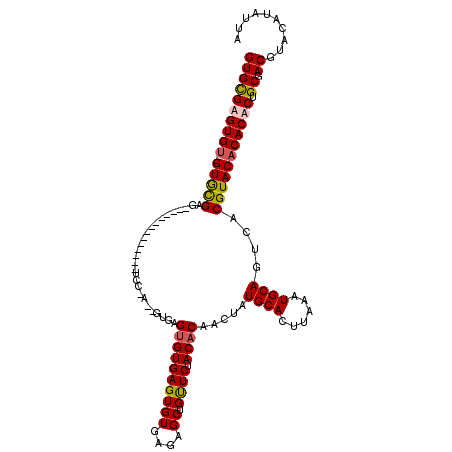
| Reading direction | forward |
| Mean pairwise identity | 80.30 |
| Mean single sequence MFE | -30.65 |
| Consensus MFE | -21.49 |
| Energy contribution | -20.93 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.70 |
| SVM decision value | 2.33 |
| SVM RNA-class probability | 0.992408 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1352732 120 + 20766785 UAAUAUGUACGUCGCAGUGUGUGUACGUGACUGCAUUUAAGUGCAUAGUUGUGUAGAACAGCUCUCACACUCACACGCACACUCGGAAUCGCACUCACACAUUCGUACUAGCACUCGCAC ......((((..((.(((((((((..((((.(((((....))))).((((((.....)))))).......)))))))))))))))((((.(........)))))))))..((....)).. ( -32.80) >DroSim_CAF1 49051 96 + 1 UAAUAUGUACGUCGCAGUGUGUGUACGUGACUGCAUUUAAGUGCAUAGUUGUGUAGAGCAGC----ACACUCACACUCACACA------------------CUCGCAC--ACACUCGCAC .............(((((((((((..(((..(((((....)))))..((.((((.(((....----...)))))))..)).))------------------)..))))--))))).)).. ( -30.70) >DroEre_CAF1 51509 102 + 1 UAAUAUGUACGUCGCAGUGUGUGUACGUGACUGCAUUUAAGUGCAUAGUUGUGUAGAACAGCUCGCACACUCACGCUCAC--UAGGA--------------AUCACAC--ACACACACAC ................((((((((..((((..........((((..((((((.....)))))).))))..((.(......--..)))--------------.))))..--)))))))).. ( -27.80) >DroYak_CAF1 38771 100 + 1 UAAUAUGUACGUCGCAGUGUGUGUACGUGACUGCAUUUAAGUGCAUAGUUGUGUAGAACAGCUCUCACACUCACAC--GC--UGAAA--------------CUCACAC--ACACACACAC .....(((..((..(((((((((...((((.(((((....))))).((((((.....)))))).))))...)))))--))--))..)--------------)..))).--.......... ( -31.30) >consensus UAAUAUGUACGUCGCAGUGUGUGUACGUGACUGCAUUUAAGUGCAUAGUUGUGUAGAACAGCUCUCACACUCACACUCAC__U_GGA______________CUCACAC__ACACACACAC ................((((((((..((((.(((((....)))))..(((((.....)))))........................................))))....)))))))).. (-21.49 = -20.93 + -0.56)

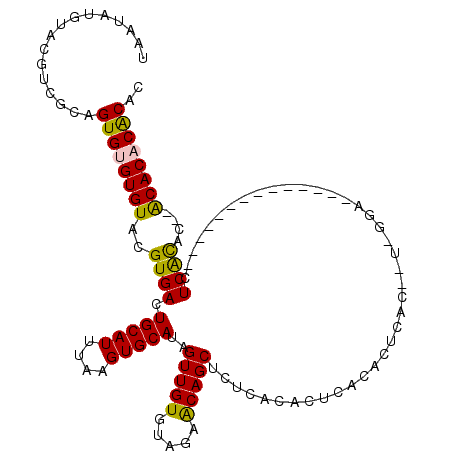
| Location | 1,352,732 – 1,352,852 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.30 |
| Mean single sequence MFE | -34.98 |
| Consensus MFE | -18.50 |
| Energy contribution | -18.62 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.719126 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1352732 120 - 20766785 GUGCGAGUGCUAGUACGAAUGUGUGAGUGCGAUUCCGAGUGUGCGUGUGAGUGUGAGAGCUGUUCUACACAACUAUGCACUUAAAUGCAGUCACGUACACACACUGCGACGUACAUAUUA (((((.(((....)))(...(((((.(((((....)..((((((((.(((((((...((.(((.....))).))..))))))).)))))..))))))).)))))..)..)))))...... ( -36.40) >DroSim_CAF1 49051 96 - 1 GUGCGAGUGU--GUGCGAG------------------UGUGUGAGUGUGAGUGU----GCUGCUCUACACAACUAUGCACUUAAAUGCAGUCACGUACACACACUGCGACGUACAUAUUA .....(((((--(((((((------------------((((((..(((((((((----(......))))).....((((......)))).)))))..))))))))....)))))))))). ( -36.50) >DroEre_CAF1 51509 102 - 1 GUGUGUGUGU--GUGUGAU--------------UCCUA--GUGAGCGUGAGUGUGCGAGCUGUUCUACACAACUAUGCACUUAAAUGCAGUCACGUACACACACUGCGACGUACAUAUUA ((((((((((--(((((((--------------(.(..--..).((((....(((((((.(((.....))).)).)))))....)))))))))))))))))))).))............. ( -33.10) >DroYak_CAF1 38771 100 - 1 GUGUGUGUGU--GUGUGAG--------------UUUCA--GC--GUGUGAGUGUGAGAGCUGUUCUACACAACUAUGCACUUAAAUGCAGUCACGUACACACACUGCGACGUACAUAUUA ((((((((((--((((((.--------------(....--((--((.(((((((...((.(((.....))).))..))))))).))))).)))))))))))))).))............. ( -33.90) >consensus GUGCGAGUGU__GUGCGAG______________UCC_A__GUGAGUGUGAGUGUGAGAGCUGUUCUACACAACUAUGCACUUAAAUGCAGUCACGUACACACACUGCGACGUACAUAUUA ((((((((((..(((((...........................((((((((((....)).)))).)))).....((((......))))....))))))))))).)).)).......... (-18.50 = -18.62 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:27:25 2006