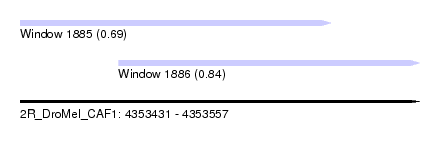
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,353,431 – 4,353,557 |
| Length | 126 |
| Max. P | 0.838895 |

| Location | 4,353,431 – 4,353,529 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 84.38 |
| Mean single sequence MFE | -27.40 |
| Consensus MFE | -21.40 |
| Energy contribution | -22.07 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.691264 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4353431 98 + 20766785 UAUGCAAUUUGUGAAUGG----GCACAAAGGCAAC--GAAAAGGGGAAAGGAAAGGGAGCCG-----GCAGUGGAAAAUCGUCUGGCAUUUUAGGCAUUUUCCAUUGCU ..(((..((((((.....----.)))))).)))..--............((........))(-----((((((((((((.((((((....))))))))))))))))))) ( -30.10) >DroSec_CAF1 23987 98 + 1 UAUGCAAUUUGUGAAUGG----GCACAAAGGCAAC--GAAAAGGGGAAAGGAAAGGGAGCCG-----GCAGUGGAAAAUCGUCUGGCAUUUUAGGCAUUUUCCAUUGCU ..(((..((((((.....----.)))))).)))..--............((........))(-----((((((((((((.((((((....))))))))))))))))))) ( -30.10) >DroSim_CAF1 24270 98 + 1 UAUGCAAUUUGUGAAUGG----GCACAAAGGCAAC--GAAGAGGGGAAGGGAAAGGGAGCGG-----GCAGUGGAAAAUCGUCUGGCAUUUUAGGCAUUUUCCAUUGCU ..(((..((((((.....----.))))))(....)--.....................)))(-----((((((((((((.((((((....))))))))))))))))))) ( -30.20) >DroEre_CAF1 21127 101 + 1 UAUGCAAUUUGUGAAUGGGCGGGCACAAAAGCAAC--GAAAAGGGGCAG-GAAGAGGAGAAG-----GCUGUGGAAAAUCGUCUGGCGUUUUAGGCAUUUUCCAUUGCU .......((((((..(....)..)))))).((..(--.....)..))..-...........(-----((.(((((((((.((((((....))))))))))))))).))) ( -26.10) >DroYak_CAF1 22300 95 + 1 UAUGCAAUUUGUGAAUGG----GCACAAAAGCGAC--GAAAAGGGGUAG-AAAGG--GGCCG-----GCAGUGGAAAAUCGUCUGGCAUUUUAGGCAUUUUCCAUUGCU ..(((..((((((.....----.)))))).)))..--.......(((..-.....--.)))(-----((((((((((((.((((((....))))))))))))))))))) ( -30.30) >DroAna_CAF1 30753 104 + 1 UAUGCAAUUUCUGAAUGG----GCACAAAAAUGAAGGGAAAAAAAGGACACAAAGCGAGCAGGAGCAGAAGCAGGAAAUCGUCUGGCAUUUUAGGCAUUUU-CAUUGCU ...((((((((((.....----((.((....))............(....).......)).....)))))...((((((.((((((....)))))))))))-)))))). ( -17.60) >consensus UAUGCAAUUUGUGAAUGG____GCACAAAAGCAAC__GAAAAGGGGAAGGGAAAGGGAGCCG_____GCAGUGGAAAAUCGUCUGGCAUUUUAGGCAUUUUCCAUUGCU ..(((..((((((..........)))))).)))..................................((((((((((((.((((((....)))))))))))))))))). (-21.40 = -22.07 + 0.67)

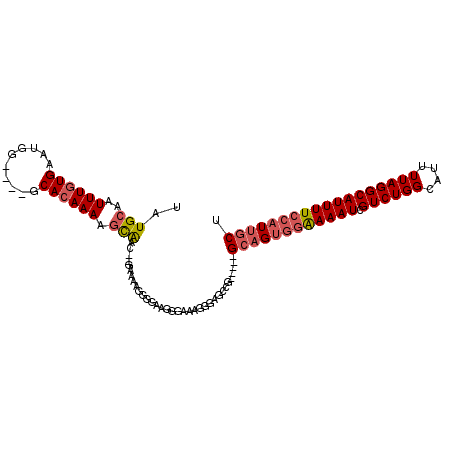
| Location | 4,353,462 – 4,353,557 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 83.84 |
| Mean single sequence MFE | -32.48 |
| Consensus MFE | -21.87 |
| Energy contribution | -24.07 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.37 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.838895 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4353462 95 + 20766785 -GAAAAGGGGAAAGGAAAGGGAGCCG-----GCAGUGGAAAAUCGUCUGGCAUUUUAGGCAUUUUCCAUUGCUAAAUAGAUGUCCUGCAGGCGCAGGGGCG -.....................((((-----((((((((((((.((((((....)))))))))))))))))))..........(((((....)))))))). ( -36.10) >DroSec_CAF1 24018 95 + 1 -GAAAAGGGGAAAGGAAAGGGAGCCG-----GCAGUGGAAAAUCGUCUGGCAUUUUAGGCAUUUUCCAUUGCUUAAUAGAUGUCCUGCAGGCGCAGGGGCG -.....................((((-----((((((((((((.((((((....)))))))))))))))))))..........(((((....)))))))). ( -35.50) >DroSim_CAF1 24301 95 + 1 -GAAGAGGGGAAGGGAAAGGGAGCGG-----GCAGUGGAAAAUCGUCUGGCAUUUUAGGCAUUUUCCAUUGCUAAAUAGAUGUCCUGCAGGCGCAGGGGCG -.....................((.(-----((((((((((((.((((((....))))))))))))))))))).........((((((....)))))))). ( -33.50) >DroEre_CAF1 21162 85 + 1 -GAAAAGGGGCAG-GAAGAGGAGAAG-----GCUGUGGAAAAUCGUCUGGCGUUUUAGGCAUUUUCCAUUGCUAAAUAGAUGUCCUGCAGGC--------- -........((((-((.........(-----((.(((((((((.((((((....))))))))))))))).))).........))))))....--------- ( -29.47) >DroYak_CAF1 22331 92 + 1 -GAAAAGGGGUAG-AAAGG--GGCCG-----GCAGUGGAAAAUCGUCUGGCAUUUUAGGCAUUUUCCAUUGCUAAAUAGAUGUCCUGCAGGGGCAGGGGCG -............-.....--.((((-----((((((((((((.((((((....)))))))))))))))))))..........(((((....)))))))). ( -35.80) >DroAna_CAF1 30785 100 + 1 GGAAAAAAAGGACACAAAGCGAGCAGGAGCAGAAGCAGGAAAUCGUCUGGCAUUUUAGGCAUUUU-CAUUGCUAAAUAGAUGUCCUGCAGGCACAGGGGCG ..................((..((((((.((..(((((((((..((((((....))))))..)))-).))))).......))))))))..))......... ( -24.50) >consensus _GAAAAGGGGAAGGGAAAGGGAGCCG_____GCAGUGGAAAAUCGUCUGGCAUUUUAGGCAUUUUCCAUUGCUAAAUAGAUGUCCUGCAGGCGCAGGGGCG ......................((.......((((((((((((.((((((....))))))))))))))))))..........((((((....)))))))). (-21.87 = -24.07 + 2.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:36 2006