
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,352,036 – 4,352,138 |
| Length | 102 |
| Max. P | 0.681195 |

| Location | 4,352,036 – 4,352,138 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 77.65 |
| Mean single sequence MFE | -37.77 |
| Consensus MFE | -20.65 |
| Energy contribution | -22.66 |
| Covariance contribution | 2.01 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.681195 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
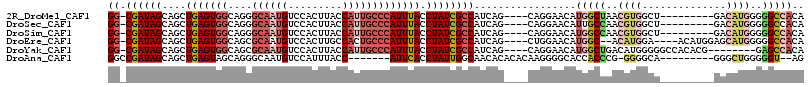
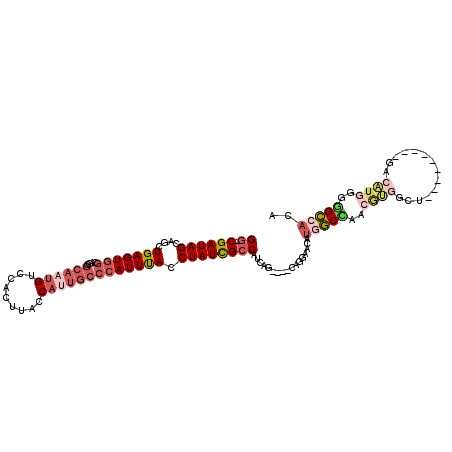
>2R_DroMel_CAF1 4352036 102 + 20766785 GG-CGAUAGCAGCUGAGUGGCAGGGCAAUGUCCACUUACCAUUGCCCAUUUACCUAUCGCCAUCAG----CAGGAACAUGGCUAACGUGGCU---------GACAUGGGGGCCACA ((-((((((..(((....))).((((((((.........))))))))......)))))))).....----........(((((..((((...---------..))))..))))).. ( -42.80) >DroSec_CAF1 22603 102 + 1 GG-CGAUAGCAGCUGAGUGGCAGGGCAAUGUCCACUUACCAUUGCCCAUUUACCUAUCGCCAUCAG----CAGGAACAUUGCCAACGUGGCU---------GACAUGGGGGCCACA ((-((((((..(((....))).((((((((.........))))))))......))))))))....(----(((.....))))....((((((---------........)))))). ( -41.20) >DroSim_CAF1 22877 102 + 1 GG-CGAUAGCAGCUGAGUGGCAGGGCAAUGUCCACUUACCAUUGCCCAUUUACCUAUCGCCAUCAG----CAGGAACAUGGCCAACGUGGCU---------GACAUGGGGGCCACA ((-((((((..(((....))).((((((((.........))))))))......)))))))).....----........(((((..((((...---------..))))..))))).. ( -44.70) >DroEre_CAF1 19731 105 + 1 GG-CGAUAGCAGCUGAGUGGCAGCGCAAUGUCCACUUGCCACUGCCCAUUUACCUAUCGCCAUCAG----CUGGAACAUGGC--ACAUGGA----ACAUGGAGCAUGGGGGCCACA ((-((((((....((((((((((.((((.(....)))))..))).))))))).))))))))....(----.(((..(...((--.((((..----.))))..))....)..)))). ( -35.30) >DroYak_CAF1 20892 103 + 1 GG-CGAUAGCAGCUGAGUGGCAGCGCAAUGUCCACUUACCAUUGCCCAUUUACCUAUCGCCAUCAG----CAGGAACAUGGCUGACAUGGGGGCCACACG--------GAGCCACA ((-((((((..((((.....))))((((((.........))))))........)))))))).((((----(.........)))))....((..((....)--------)..))... ( -34.00) >DroAna_CAF1 29564 97 + 1 GGCCGAUAGCAGCUGAGUAGCAGGGCAAUGUCCAUUUACC-------AUUCACCUAUUGGCAACACACACAAGGGGCACCACCCG-GGGGCA---------GGGCUGGGGGU--AG .((((((((..(((....))).((((...)))).......-------......))))))))................(((.((((-(.....---------...))))))))--.. ( -28.60) >consensus GG_CGAUAGCAGCUGAGUGGCAGGGCAAUGUCCACUUACCAUUGCCCAUUUACCUAUCGCCAUCAG____CAGGAACAUGGCCAACGUGGCU_________GACAUGGGGGCCACA ((.((((((....(((((((....((((((.........))))))))))))).)))))))).................(((((..((((..............))))..))))).. (-20.65 = -22.66 + 2.01)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:34 2006