
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
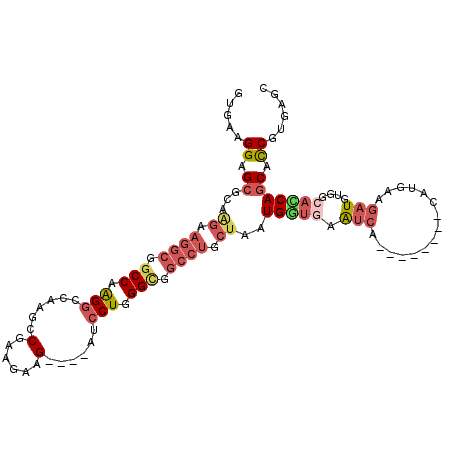
| Location | 4,338,411 – 4,338,511 |
| Length | 100 |
| Max. P | 0.618045 |

| Location | 4,338,411 – 4,338,511 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 78.85 |
| Mean single sequence MFE | -37.00 |
| Consensus MFE | -16.71 |
| Energy contribution | -18.04 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.618045 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
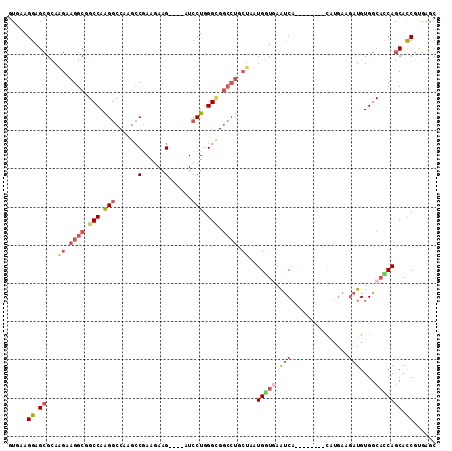
>2R_DroMel_CAF1 4338411 100 + 20766785 GUAAAGGAGCGCAAGAAGGCGGCCAAGGCCAAGCCGAAGAAG----AUCCUGGGCGGCCUGCUUAUGGUGAAUCA--------CAUGAAGAUGUGGCACCAGCACCGUGAGC .....((..(((......))).)).(((((..(((..((...----...)).))))))))((((((((((..(((--------(((....))))))......)))))))))) ( -37.70) >DroSec_CAF1 8878 100 + 1 GUAAAGGAGCGCAAGAAGGCGGCCAAGGCCAAGCCGAAGAAG----GUCCUGGGUGGCCUGCUAAUGGUGAAUCA--------UAUGAAGAUGUGGCACCAGCACCGUGAGC .....((.((...((.((((.(((.(((....(((......)----))))).))).)))).))..(((((...((--------(((....))))).))))))).))...... ( -34.80) >DroSim_CAF1 8905 100 + 1 GUUAAGGAGCGUAAGAAGGCGGCCAAGGCCAAGCCGAAAAAG----GUCCUGGGUGGCCUGCUAAUGGUGAAUCA--------UAUGAAGAUGUGGCACCAGCACCGUGAGC (((..((.((...((.((((.(((.(((....(((......)----))))).))).)))).))..(((((...((--------(((....))))).))))))).))...))) ( -35.20) >DroEre_CAF1 6448 100 + 1 GUGAAGGAGCGCAGGAAGGCGGCCAGGGCCAAGCCGAAGAAG----AUCCUGGGCGGCCUCCUGAUGCUGAACCA--------CAUGAAGAUGUGGCACCAGCAUCGGGAGC .....((.((.(((((.((((((....)))..))).......----.))))).))..))(((((((((((..(((--------(((....))))))...))))))))))).. ( -48.60) >DroYak_CAF1 7074 100 + 1 GUGAAGGAGCGCAAGAAGGCUGCCAGGGCCAAGCCGAAGAAG----AUCCUGGGCGGCCUCCUGAUGGUAAAUCA--------CUUGAAGAUGUGGCAUCAGCAUCGUGAGC ........((.((.(((((((((((((..(...........)----..))).)))))))).((((((.((.(((.--------......))).)).))))))..)).)).)) ( -32.10) >DroAna_CAF1 15784 100 + 1 GUGAAGGAGCGGAAGAAGGCUGCCAAGAC---GGCAAAAAUGUCGAUUCCUGGGA---------AUGCUCAGUCCUUAAUGCGCCACAAAAUGUGGCUGCAACACCGCGAAC ........((((...(((((((......(---((((....)))))((((....))---------))...))).))))..((((((((.....))))).)))...)))).... ( -33.60) >consensus GUGAAGGAGCGCAAGAAGGCGGCCAAGGCCAAGCCGAAGAAG____AUCCUGGGCGGCCUGCUAAUGGUGAAUCA________CAUGAAGAUGUGGCACCAGCACCGUGAGC .....((.((...((.((((.(((.(((......(......)......))).))).)))).))..(((((.(((...............)))....))))))).))...... (-16.71 = -18.04 + 1.34)

| Location | 4,338,411 – 4,338,511 |
|---|---|
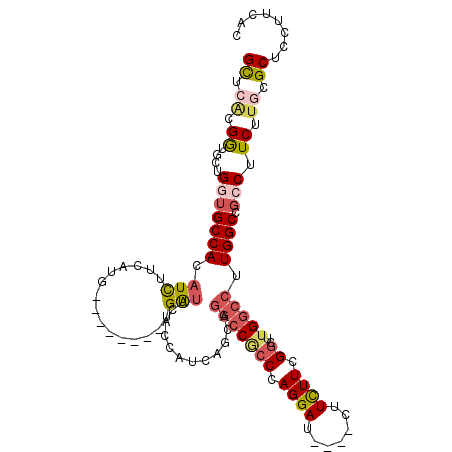
| Length | 100 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 78.85 |
| Mean single sequence MFE | -33.93 |
| Consensus MFE | -13.44 |
| Energy contribution | -14.34 |
| Covariance contribution | 0.91 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.40 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
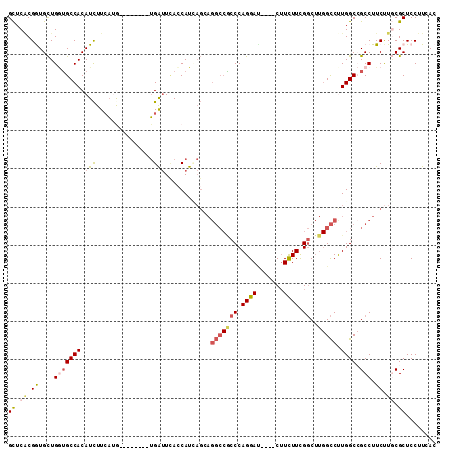
>2R_DroMel_CAF1 4338411 100 - 20766785 GCUCACGGUGCUGGUGCCACAUCUUCAUG--------UGAUUCACCAUAAGCAGGCCGCCCAGGAU----CUUCUUCGGCUUGGCCUUGGCCGCCUUCUUGCGCUCCUUUAC ......((.(((((((.(((((....)))--------))...)))))((((.((((.(((.(((..----...))).)))..(((....))))))).)))).)).))..... ( -35.10) >DroSec_CAF1 8878 100 - 1 GCUCACGGUGCUGGUGCCACAUCUUCAUA--------UGAUUCACCAUUAGCAGGCCACCCAGGAC----CUUCUUCGGCUUGGCCUUGGCCGCCUUCUUGCGCUCCUUUAC ((....((((((((((....(((......--------.)))....))))))).(((((.(((((.(----(......)))))))...)))))))).....)).......... ( -31.00) >DroSim_CAF1 8905 100 - 1 GCUCACGGUGCUGGUGCCACAUCUUCAUA--------UGAUUCACCAUUAGCAGGCCACCCAGGAC----CUUUUUCGGCUUGGCCUUGGCCGCCUUCUUACGCUCCUUAAC ......((.(((((((...(((......)--------))...)))))...((.(((((.(((((.(----(......)))))))...)))))))........)).))..... ( -29.80) >DroEre_CAF1 6448 100 - 1 GCUCCCGAUGCUGGUGCCACAUCUUCAUG--------UGGUUCAGCAUCAGGAGGCCGCCCAGGAU----CUUCUUCGGCUUGGCCCUGGCCGCCUUCCUGCGCUCCUUCAC ((....((((((((.(((((((....)))--------))))))))))))((((((.((.(((((..----(..(....)...)..))))).)).)))))))).......... ( -45.50) >DroYak_CAF1 7074 100 - 1 GCUCACGAUGCUGAUGCCACAUCUUCAAG--------UGAUUUACCAUCAGGAGGCCGCCCAGGAU----CUUCUUCGGCUUGGCCCUGGCAGCCUUCUUGCGCUCCUUCAC ((.((.((((.(((...(((........)--------))..))).))))(((((((.(((.(((..----(..(....)...)..)))))).))))))))).))........ ( -31.80) >DroAna_CAF1 15784 100 - 1 GUUCGCGGUGUUGCAGCCACAUUUUGUGGCGCAUUAAGGACUGAGCAU---------UCCCAGGAAUCGACAUUUUUGCC---GUCUUGGCAGCCUUCUUCCGCUCCUUCAC ....((((...(((.(((((.....))))))))..((((.(((.....---------..((((((..((.((....)).)---))))))))))))))...))))........ ( -30.41) >consensus GCUCACGGUGCUGGUGCCACAUCUUCAUG________UGAUUCACCAUCAGCAGGCCGCCCAGGAU____CUUCUUCGGCUUGGCCUUGGCCGCCUUCUUGCGCUCCUUCAC ((.((.((....(((((((.(((...............)))............(((((((.((((.......)))).))..))))).)))).))).)).)).))........ (-13.44 = -14.34 + 0.91)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:30 2006