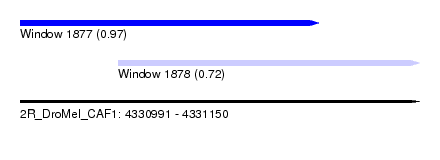
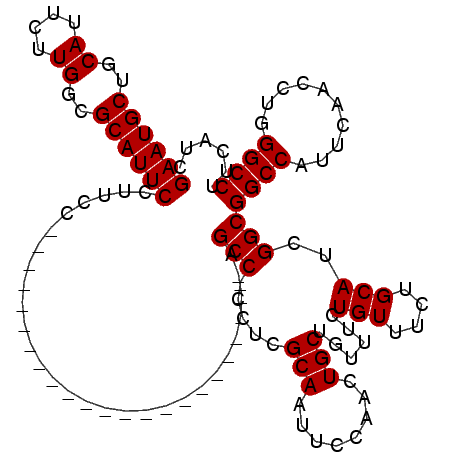
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,330,991 – 4,331,150 |
| Length | 159 |
| Max. P | 0.965889 |

| Location | 4,330,991 – 4,331,110 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.49 |
| Mean single sequence MFE | -38.40 |
| Consensus MFE | -24.86 |
| Energy contribution | -24.53 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.965889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4330991 119 + 20766785 AGUAGAUGGGCCG-UUUCAAGUGGGUGGUUAACAAGGACGGAAUGCUGCAUUCUUGGAGCAUUCCUUCCCAGUCCCCAGGCCCCAGCCUCCCAGCCACCCAGCAAUUCCAACUGCUGUUU (((((.((((..(-(......(((((((((.....(((.((((((((.((....)).)))))))).)))........((((....))))...)))))))))))...)))).))))).... ( -46.70) >DroEre_CAF1 9332 95 + 1 AGCAGAUGGGCCG-CUUGAAGUGGGUGGCUAACAAGGACGGAAUGCCGCAUUCUUGGCGCAUUCCUUCC-----------------------CGCC-CCUCGCAGUUCCAACUGCUGUUU (((((.(((((((-(((.....))))))).(((..(((.(((((((.((.......))))))))).)))-----------------------.((.-....)).)))))).))))).... ( -37.70) >DroYak_CAF1 10407 97 + 1 AGUAGACGGGCCCUUUUGAAGUGGGUGGCUAACAAGGACGGAAUGCUGCAUUCUUGGCGCAUUCCUCGC-----------------------AGCCCCCUCGCAAUUCCAACUGCUGUUU (((((..(((.....(((....(((.((((.....(((.(((((((.((.......))))))))))).)-----------------------)))))))...))).)))..))))).... ( -30.80) >consensus AGUAGAUGGGCCG_UUUGAAGUGGGUGGCUAACAAGGACGGAAUGCUGCAUUCUUGGCGCAUUCCUUCC_______________________AGCC_CCUCGCAAUUCCAACUGCUGUUU (((((...(((((.((......)).))))).....(((.(((((((..((....))..)))))))............................((......))...)))..))))).... (-24.86 = -24.53 + -0.33)

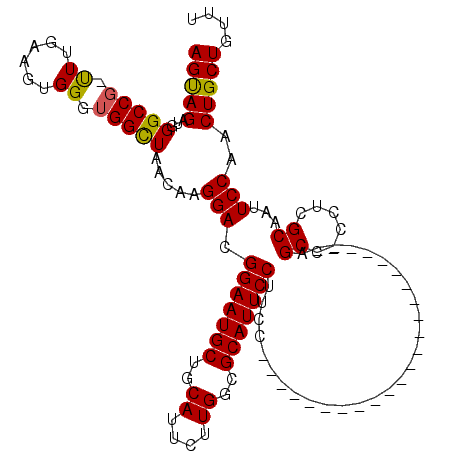
| Location | 4,331,030 – 4,331,150 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.71 |
| Mean single sequence MFE | -27.97 |
| Consensus MFE | -20.13 |
| Energy contribution | -20.13 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.718029 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4331030 120 + 20766785 GAAUGCUGCAUUCUUGGAGCAUUCCUUCCCAGUCCCCAGGCCCCAGCCUCCCAGCCACCCAGCAAUUCCAACUGCUGUUUCUGUUUCUGCAUCGGCGGCCAUUCAACCUGGGCCUUCAUC (((((((.((....)).)))))))...(((((.....((((....))))....(((.(((((((........)))))....(((....)))..)).)))........)))))........ ( -36.50) >DroEre_CAF1 9371 96 + 1 GAAUGCCGCAUUCUUGGCGCAUUCCUUCC-----------------------CGCC-CCUCGCAGUUCCAACUGCUGUUUCUGUUUCUGCAUCGGCGGCCAUUCAACCUGGGCCUUCAUC ((((((.((.......)))))))).....-----------------------.(((-....(((((....)))))......(((....)))..)))((((..........))))...... ( -25.80) >DroYak_CAF1 10447 97 + 1 GAAUGCUGCAUUCUUGGCGCAUUCCUCGC-----------------------AGCCCCCUCGCAAUUCCAACUGCUGUUUCUGUUUCUGCAUCGGCGGCCAUUCAACCUGGGCCUUCAUC ((((((.((.......)))))))).....-----------------------.(((.....(((........)))......(((....)))..)))((((..........))))...... ( -21.60) >consensus GAAUGCUGCAUUCUUGGCGCAUUCCUUCC_______________________AGCC_CCUCGCAAUUCCAACUGCUGUUUCUGUUUCUGCAUCGGCGGCCAUUCAACCUGGGCCUUCAUC ((((((..((....))..)))))).............................(((.....(((........)))......(((....)))..)))((((..........))))...... (-20.13 = -20.13 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:29 2006