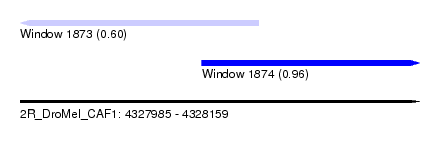
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,327,985 – 4,328,159 |
| Length | 174 |
| Max. P | 0.955470 |

| Location | 4,327,985 – 4,328,089 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.30 |
| Mean single sequence MFE | -37.13 |
| Consensus MFE | -19.87 |
| Energy contribution | -20.23 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.604481 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4327985 104 - 20766785 UCCAAG---------------GAGCUCGGGGCUGACUUUAAGGUCAGCCGCCUCACAUAACAAAUGUCGGGCGAUAUUUCC-CAGGAAUGCCGGCAGAUGAAAACUUUCCUCAUUUGCAC (((..(---------------(((.((..((((((((....))))))))((((.((((.....)))).))))))...))))-..)))......((((((((.........)))))))).. ( -39.30) >DroPse_CAF1 26630 114 - 1 CCCAAGACGGCUGCGUCCGUGGAG--UG-GACUGGCUUUAAGGUCAG---CUUCACAUAACAAAUGCCGGGCGAUAUUUCUUCGGGAAAGCUGAGAGAUGAAAACUUUCCUCAUUUGCAC .(((.((((....))))..))).(--((-((((((((....))))))---.)))))....((((((...(((....((((....)))).)))(((((.(....))))))..))))))... ( -32.80) >DroSec_CAF1 7232 104 - 1 CCCAAG---------------GAGCUCAGGGCUGACUUUAAGGUCAGCCGCCUCACAUAACAAAUGUCGGGCGAUAUUUCC-CGGGAAUGCCGGCAGAUGAAAACUUUCCUCAUUUGCAC (((..(---------------(((.((..((((((((....))))))))((((.((((.....)))).))))))...))))-.))).......((((((((.........)))))))).. ( -39.90) >DroPer_CAF1 26469 114 - 1 CCCAAGACGGCUGGGUCCGUGGAG--UG-GACUGGCUUUAAGGUCAG---CUUCACAUAACAAAUGCCGGGCGAUAUUUCUUGGGGAAAGCUGAGAGAUGAAAACUUUCCUCAUUUGCAC (((((((((.((((.((....))(--((-((((((((....))))))---.)))))..........)))).)).....)))))))....((((((.((.........))))))...)).. ( -36.80) >DroEre_CAF1 6503 100 - 1 CCCAAG---------------GAGCUCG-GGCUGACUUUAAGGUCAG---CCUCACAUAACAAAUGUGGGGCGAUAUUUCC-CGGGAAUGCCGGCAGAUGAAAACUUUCCUCAUUUGCAC .....(---------------(..((((-((.(((((....)))))(---((((((((.....))))))))).......))-))))....)).((((((((.........)))))))).. ( -40.50) >DroYak_CAF1 7358 100 - 1 CCCUUG---------------GAGCUCG-GGCUGACUUUAAGGUCAG---CCUCACAUAACAAAUGUCGGGCGAUAUUUCC-CGGGAAUGCCGGCAGAUAAAAACUUUCCUCAUUUGCAC .....(---------------(((((((-((((((((....))))))---))..((((.....)))))))))..(((((.(-(((.....)))).))))).......))).......... ( -33.50) >consensus CCCAAG_______________GAGCUCG_GGCUGACUUUAAGGUCAG___CCUCACAUAACAAAUGUCGGGCGAUAUUUCC_CGGGAAUGCCGGCAGAUGAAAACUUUCCUCAUUUGCAC (((..................(((.......((((((....))))))....))).(((.....)))..)))((.((((((....)))))).))((((((((.........)))))))).. (-19.87 = -20.23 + 0.36)
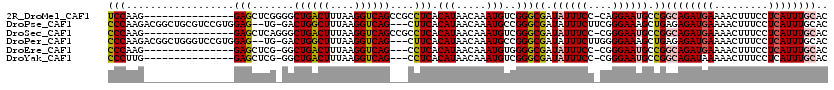
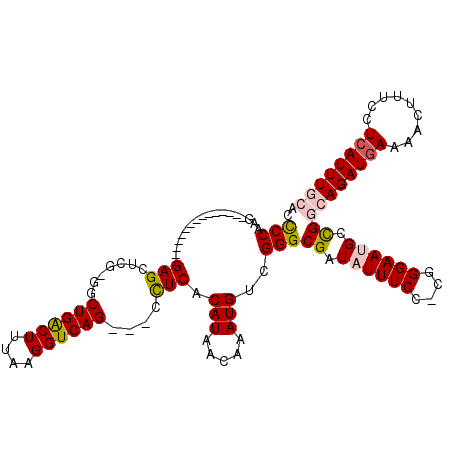
| Location | 4,328,064 – 4,328,159 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.14 |
| Mean single sequence MFE | -33.18 |
| Consensus MFE | -13.21 |
| Energy contribution | -16.61 |
| Covariance contribution | 3.40 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.40 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.955470 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4328064 95 + 20766785 UAAAGUCAGCCCCGAGCUC---------------CUUGGA--UCGCACUCCUCGUGUGACCGGGCUC-CUGAUGGCUCAACUUACAUCACAGCAAGCCAUCCGGCCAGCUCCC------- ...(((..(((..((((((---------------...((.--((((((.....))))))))))))))-..(((((((...((........))..))))))).)))..)))...------- ( -32.50) >DroPse_CAF1 26707 115 + 1 UAAAGCCAGUC-CA--CUCCACGGACGCAGCCGUCUUGGGGCCCGCACUUCGGGUGUGACUGUGUCCCGCAAUGGCUCAAGCAGCAUU--GGCUAGCCACCCGCCCUUCCGCCACCCGCC ...((((((((-(.--......))))((((((((..((((((((((((.....)))))...).))))))..))))))...)).....)--)))).((.....((......)).....)). ( -37.50) >DroSec_CAF1 7311 95 + 1 UAAAGUCAGCCCUGAGCUC---------------CUUGGG--UCGCACUCCUCGUGUGACCGGGCUC-CUGAUGGCUCAACUUACAUCACAGCCAGCCAUCCGGCCAGCUCCC------- ...(((..(((..((((((---------------....((--((((((.....))))))))))))))-..(((((((...((........))..))))))).)))..)))...------- ( -35.60) >DroEre_CAF1 6579 81 + 1 UAAAGUCAGCC-CGAGCUC---------------CUUGGG--UCGCACUCCUCGUGUGACCGGACUC-CCGAUGGCUCAACUUACAUC-------------CGACCAGCUCCC------- ...........-.(((((.---------------.(((((--((((((.....))))))))((....-))((((..........))))-------------)))..)))))..------- ( -25.30) >DroYak_CAF1 7434 94 + 1 UAAAGUCAGCC-CGAGCUC---------------CAAGGG--UCGCACUCCUCGUGUGACCAGGCUC-CUGAUGGCUCAACUUACAUCACAGGCAGCCAUUCGGCCAGCUCCC------- ...........-.((((((---------------(...((--((((((.....)))))))).(((((-((((((..........))))..))).))))....))..)))))..------- ( -35.00) >consensus UAAAGUCAGCC_CGAGCUC_______________CUUGGG__UCGCACUCCUCGUGUGACCGGGCUC_CUGAUGGCUCAACUUACAUCACAGCCAGCCAUCCGGCCAGCUCCC_______ .............(((((...................((...((((((.....)))))))).((((....(((((((.................))))))).)))))))))......... (-13.21 = -16.61 + 3.40)

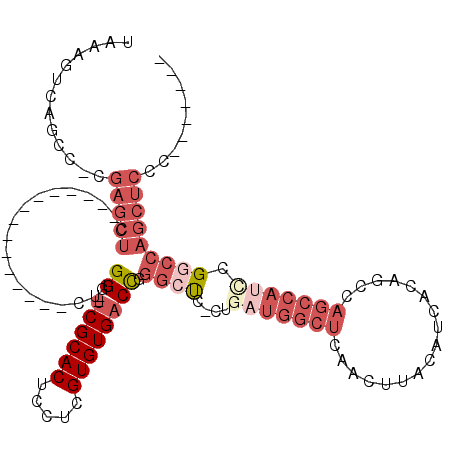
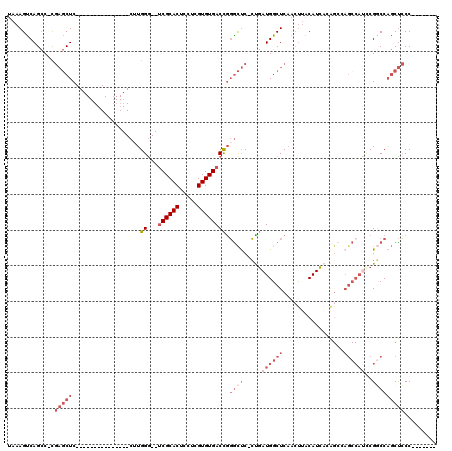
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:25 2006