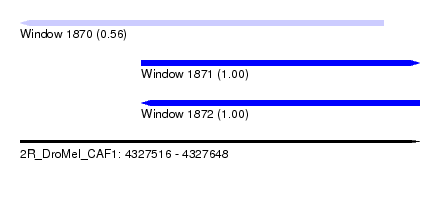
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,327,516 – 4,327,648 |
| Length | 132 |
| Max. P | 0.998989 |

| Location | 4,327,516 – 4,327,636 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.00 |
| Mean single sequence MFE | -26.30 |
| Consensus MFE | -25.26 |
| Energy contribution | -25.10 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.559940 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4327516 120 - 20766785 UUUGUACUCCAAAUUAAUGGCAUGCGAAAAUUUUUGGGGAAUUUCCAUAAAUUUGCACGUGGACUUUUGUGUGCAAAAGUUCUUUGCUUUGCCAAACUUAUAAUCUAAUUGCGCCCUCAC .................(((((.((((((((((.((((.....)))).))))))......(((((((((....))))))))).))))..))))).......................... ( -26.20) >DroSec_CAF1 6761 120 - 1 UUUGUACUCCAAAUUAAUGGCAUGCGAAAAUUUUUGGGGAAUUUCCAUAAAUUUGCACGUGGACUUUUGUGUGCAAAAGUUCUUUGCUUUGCCAAACUUAUAAUCUAAUUGCGCCCUCAC .................(((((.((((((((((.((((.....)))).))))))......(((((((((....))))))))).))))..))))).......................... ( -26.20) >DroSim_CAF1 8743 120 - 1 UUUGUACUCCAAAUUAAUGGCAUGCGAAAAUUUUUGGGGAAUUUCCAUAAAUUUGCACGUGGACUUUUGUGUGCAAAAGUUCUUUGCUUUGCCAAACUUAUAAUCUAAUUGCGCCCUCAC .................(((((.((((((((((.((((.....)))).))))))......(((((((((....))))))))).))))..))))).......................... ( -26.20) >DroEre_CAF1 6028 120 - 1 UUUGUACUCCAAAUUAAUGGCAUGCGAACAUUUUUGGGGAAUUUCCAUAAAUUUGCACGUGGACUUUUGUGUGCGAAAGUUCUUUGCAUUGCCAAACUUAUAAUCUAAUUGCGCCCUCAC .................(((((((((((((..((((.((.....)).))))..)).....(((((((((....))))))))))))))).))))).......................... ( -26.70) >DroYak_CAF1 6902 120 - 1 UUUGUACUCCAAAUUAAUGGCAUGCGAAAAUUUUUGGGGAAUUUCCAUAAAUUUGCACGUGGACUUUUGUGUGCAAAAGUUCUUUGCUUUGCCAAACUUAUAAUCUAAUUGCGCCCUCAC .................(((((.((((((((((.((((.....)))).))))))......(((((((((....))))))))).))))..))))).......................... ( -26.20) >consensus UUUGUACUCCAAAUUAAUGGCAUGCGAAAAUUUUUGGGGAAUUUCCAUAAAUUUGCACGUGGACUUUUGUGUGCAAAAGUUCUUUGCUUUGCCAAACUUAUAAUCUAAUUGCGCCCUCAC ((((((..(((......)))..))))))......(((((...............(((...(((((((((....)))))))))..)))...(((((.............))).))))))). (-25.26 = -25.10 + -0.16)

| Location | 4,327,556 – 4,327,648 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 96.54 |
| Mean single sequence MFE | -20.50 |
| Consensus MFE | -17.12 |
| Energy contribution | -17.48 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.61 |
| SVM RNA-class probability | 0.995779 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4327556 92 + 20766785 ACUUUUGCACACAAAAGUCCACGUGCAAAUUUAUGGAAAUUCCCCAAAAAUUUUCGCAUGCCAUUAAUUUGGAGUACAAAAAGUGGAGCAAA- .((((((....))))))((((((((.((((((.(((.......))).)))))).)))...(((......)))..........))))).....- ( -22.50) >DroSec_CAF1 6801 92 + 1 ACUUUUGCACACAAAAGUCCACGUGCAAAUUUAUGGAAAUUCCCCAAAAAUUUUCGCAUGCCAUUAAUUUGGAGUACAAAAAGUGGAGCAAA- .((((((....))))))((((((((.((((((.(((.......))).)))))).)))...(((......)))..........))))).....- ( -22.50) >DroSim_CAF1 8783 93 + 1 ACUUUUGCACACAAAAGUCCACGUGCAAAUUUAUGGAAAUUCCCCAAAAAUUUUCGCAUGCCAUUAAUUUGGAGUACAAAAAGUGGAGCAAAA .((((((....))))))((((((((.((((((.(((.......))).)))))).)))...(((......)))..........)))))...... ( -22.50) >DroEre_CAF1 6068 91 + 1 ACUUUCGCACACAAAAGUCCACGUGCAAAUUUAUGGAAAUUCCCCAAAAAUGUUCGCAUGCCAUUAAUUUGGAGUACAAAAAAUG-AACAAA- ................((.(((((((..((((.(((.......))).))))....)))))(((......)))...........))-.))...- ( -12.00) >DroYak_CAF1 6942 92 + 1 ACUUUUGCACACAAAAGUCCACGUGCAAAUUUAUGGAAAUUCCCCAAAAAUUUUCGCAUGCCAUUAAUUUGGAGUACAAAAAAUGGCACAAA- (((((((....)))))))....(((.((((((.(((.......))).)))))).))).(((((((..((((.....)))).)))))))....- ( -23.00) >consensus ACUUUUGCACACAAAAGUCCACGUGCAAAUUUAUGGAAAUUCCCCAAAAAUUUUCGCAUGCCAUUAAUUUGGAGUACAAAAAGUGGAGCAAA_ (((((((....)))))))...(((((((((((.(((.......))).))))))..)))))(((((..((((.....)))).)))))....... (-17.12 = -17.48 + 0.36)

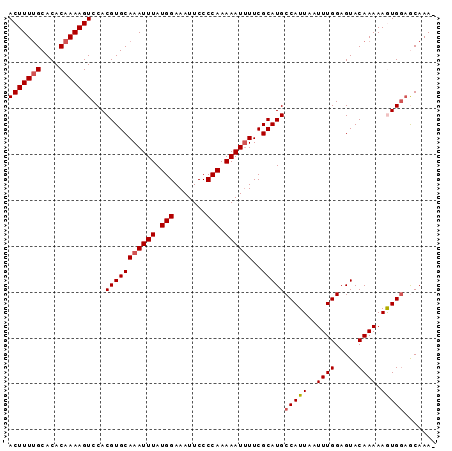
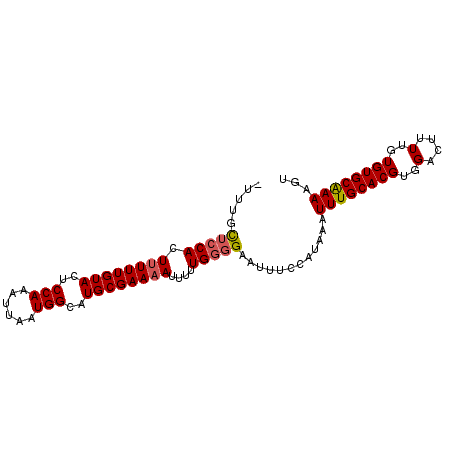
| Location | 4,327,556 – 4,327,648 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 96.54 |
| Mean single sequence MFE | -23.06 |
| Consensus MFE | -21.52 |
| Energy contribution | -21.72 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.31 |
| SVM RNA-class probability | 0.998989 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4327556 92 - 20766785 -UUUGCUCCACUUUUUGUACUCCAAAUUAAUGGCAUGCGAAAAUUUUUGGGGAAUUUCCAUAAAUUUGCACGUGGACUUUUGUGUGCAAAAGU -....(((((.((((((((..(((......)))..))))))))....)))))............((((((((..(....)..))))))))... ( -23.90) >DroSec_CAF1 6801 92 - 1 -UUUGCUCCACUUUUUGUACUCCAAAUUAAUGGCAUGCGAAAAUUUUUGGGGAAUUUCCAUAAAUUUGCACGUGGACUUUUGUGUGCAAAAGU -....(((((.((((((((..(((......)))..))))))))....)))))............((((((((..(....)..))))))))... ( -23.90) >DroSim_CAF1 8783 93 - 1 UUUUGCUCCACUUUUUGUACUCCAAAUUAAUGGCAUGCGAAAAUUUUUGGGGAAUUUCCAUAAAUUUGCACGUGGACUUUUGUGUGCAAAAGU .....(((((.((((((((..(((......)))..))))))))....)))))............((((((((..(....)..))))))))... ( -23.90) >DroEre_CAF1 6068 91 - 1 -UUUGUU-CAUUUUUUGUACUCCAAAUUAAUGGCAUGCGAACAUUUUUGGGGAAUUUCCAUAAAUUUGCACGUGGACUUUUGUGUGCGAAAGU -......-...........(((((((..((((.........)))))))))))............((((((((..(....)..))))))))... ( -19.20) >DroYak_CAF1 6942 92 - 1 -UUUGUGCCAUUUUUUGUACUCCAAAUUAAUGGCAUGCGAAAAUUUUUGGGGAAUUUCCAUAAAUUUGCACGUGGACUUUUGUGUGCAAAAGU -...((((((((.((((.....))))..))))))))(((.((((((.((((.....)))).))))))...)))..(((((((....))))))) ( -24.40) >consensus _UUUGCUCCACUUUUUGUACUCCAAAUUAAUGGCAUGCGAAAAUUUUUGGGGAAUUUCCAUAAAUUUGCACGUGGACUUUUGUGUGCAAAAGU .....(((((.((((((((..(((......)))..))))))))....)))))............((((((((..(....)..))))))))... (-21.52 = -21.72 + 0.20)

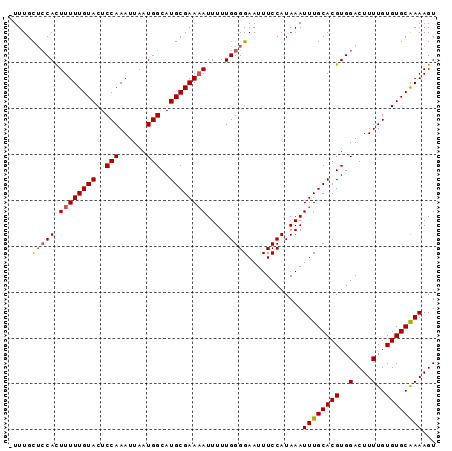
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:23 2006