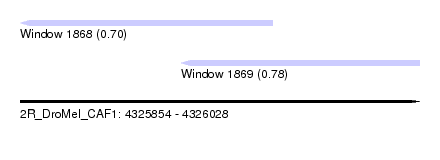
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,325,854 – 4,326,028 |
| Length | 174 |
| Max. P | 0.780961 |

| Location | 4,325,854 – 4,325,964 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 67.82 |
| Mean single sequence MFE | -33.10 |
| Consensus MFE | -9.64 |
| Energy contribution | -12.02 |
| Covariance contribution | 2.38 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.29 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.698200 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
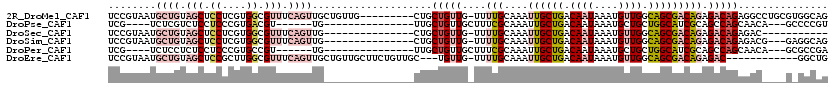
>2R_DroMel_CAF1 4325854 110 - 20766785 UCCGUAAUGCUGUAGCUCCUCGUGGCGUUUCAGUUGCUGUUG---------CUGCUGUUG-UUUUGCAAAUUGCUGACAAUAAAUGUUGGCAGCGACAGAGACAGAGGCCUGCGUGGCAG ........((....)).((.((..(.(((((.((..((((((---------(((((((((-((..((.....)).)))))).......)))))))))))..)).))))))..)).))... ( -40.61) >DroPse_CAF1 23794 92 - 1 UCG----UCUCGUCUCCUCCCGUGACGU------UG---------------UUGCUGUUGCUUUCGCAAAUUGCUGACAAUAAAUGCUGCUGGCAUCGCAGCCAGCAACA---GCCCCGU ..(----((.((........)).)))((------((---------------((((((((((....))))................(((((.......)))))))))))))---))..... ( -30.10) >DroSec_CAF1 5120 93 - 1 UCCGUAAUGCUGUAGCUCCUCGUGGCGUUUCAGUUG---------------CUGCUGUUG-UUUUGCAAAUUGCUGACAAUAAAUGUUGGCAGCGACAGAGACAGAGAC----------- ........((..(........)..))(((((.((((---------------(((((((((-((..((.....)).)))))).......))))))))).)))))......----------- ( -28.21) >DroSim_CAF1 6158 101 - 1 UCCGUAAUGCUGUAGCUCCUCGUGGCGUUUCAGUUG---------------CUGCUGUUG-UUUUGCAAAUUGCUGACAAUAAAUGUUGGCAGCGACAGAGACAGAGACG---GAGGCAG (((((...((..(........)..))(((((.((((---------------(((((((((-((..((.....)).)))))).......))))))))).)))))....)))---))..... ( -34.81) >DroPer_CAF1 23663 92 - 1 UCG----UCUCCUCUCCUCCCGUGCCGU------UG---------------UUGCUGUUGCUUUCGCAAAUUGCUGACAAUAAAUGCUGCUGGCAUCGCAGCCAGCAACA---GCGCCGA ...----.............((.((.((------((---------------((((((((((....))))................(((((.......)))))))))))))---)))))). ( -29.60) >DroEre_CAF1 4521 104 - 1 UCCGUAAUGCUGUAGCUCCGCUUGGCGUUUCAGUUGCUGUUGCUUCUGUUGC---UGUUG-UUUUGCAAAUUGCUGACAAUAAAUGUUGGCAGCGACAGAGAC------------GGCUG ........((((.(((.((....)).))).)))).((((((...((((((((---(.(((-.....)))...((..(((.....)))..))))))))))))))------------))).. ( -35.30) >consensus UCCGUAAUGCUGUAGCUCCUCGUGGCGUUUCAGUUG_______________CUGCUGUUG_UUUUGCAAAUUGCUGACAAUAAAUGUUGGCAGCGACAGAGACAGAGAC____G_GGCAG ........((((.(((.((....)).))).))))....................(((((....(((....((((((.((((....)))).))))))))).)))))............... ( -9.64 = -12.02 + 2.38)
| Location | 4,325,924 – 4,326,028 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.84 |
| Mean single sequence MFE | -31.58 |
| Consensus MFE | -19.66 |
| Energy contribution | -21.55 |
| Covariance contribution | 1.89 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.780961 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
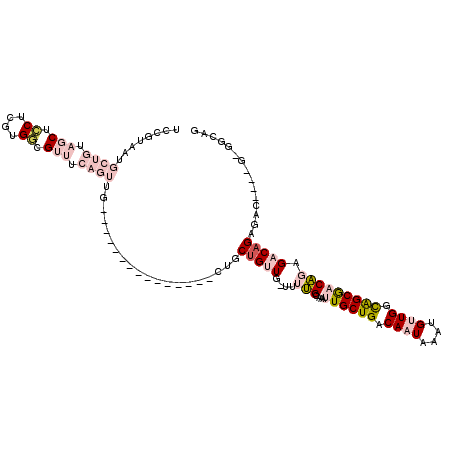
>2R_DroMel_CAF1 4325924 104 - 20766785 ------CAGUGCUGGU------UUUCCGCCC----ACUUGGGCUCCUUGGAGCAAUAAACUGCGCUAAAUUCAGCUCAGCUCCGUAAUGCUGUAGCUCCUCGUGGCGUUUCAGUUGCUGU ------(((((((((.------...))((((----....)))).......)))....((((((((((.....((((((((........)))).)))).....)))))...))))))))). ( -35.80) >DroPse_CAF1 23860 106 - 1 UCCGUACAGUUCUCGUCUUCCCUUUCCUCCUUGAUUCGUUAGCUCCUUGGAGCAAUAAACUGCGCUAAAUUCCAUUCCUCUCG----UCUCGUCUCCUCCCGUGACGU------UG---- ...(..(((((...(((...............)))......((((....))))....)))))..)................((----((.((........)).)))).------..---- ( -17.66) >DroSec_CAF1 5177 100 - 1 ------CACUGCUGGU------UUUCCGCCC----ACUUGGGCUCCUUGGAGCAAUAAACUGCGCUAAAUUCAGCUCAGCUCCGUAAUGCUGUAGCUCCUCGUGGCGUUUCAGUUG---- ------...((((((.------...))((((----....)))).......))))...((((((((((.....((((((((........)))).)))).....)))))...))))).---- ( -33.50) >DroSim_CAF1 6223 100 - 1 ------CACUGCUGGU------UUUCCGCCC----ACUUGGGCUCCUUGGAGCAAUAAACUGCGCUAAAUUCAGCUCAGCUCCGUAAUGCUGUAGCUCCUCGUGGCGUUUCAGUUG---- ------...((((((.------...))((((----....)))).......))))...((((((((((.....((((((((........)))).)))).....)))))...))))).---- ( -33.50) >DroEre_CAF1 4585 104 - 1 ------CACUCCUGGU------UUUCCGCCC----ACUUGGGCUCCUUGGAGCAAUAAACUGCGCUAAAUUCAGCUCAGCUCCGUAAUGCUGUAGCUCCGCUUGGCGUUUCAGUUGCUGU ------...(((.((.------...))((((----....)))).....)))(((((.....(((((((....((((((((........)))).))))....)))))))....)))))... ( -35.70) >DroYak_CAF1 5250 104 - 1 ------CACUCCUGGU------UUUCCGCCC----ACUUGGGCUCCUUGGAGCAAUAAACUGCGCUAAAUUCAGUUCAGCUCCGUAAUGCUGUAGCUCCGCUUGGCGUUUCAGUUGCUGU ------...(((.((.------...))((((----....)))).....)))(((((.....(((((((....((((((((........)))).))))....)))))))....)))))... ( -33.30) >consensus ______CACUGCUGGU______UUUCCGCCC____ACUUGGGCUCCUUGGAGCAAUAAACUGCGCUAAAUUCAGCUCAGCUCCGUAAUGCUGUAGCUCCUCGUGGCGUUUCAGUUG____ .........................................((((....))))....((((((((((.....((((((((........)))).)))).....)))))...)))))..... (-19.66 = -21.55 + 1.89)

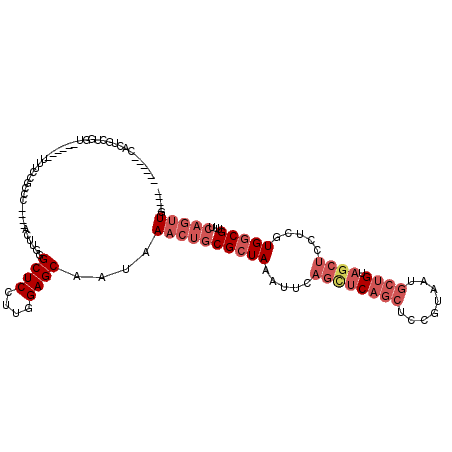
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:21 2006