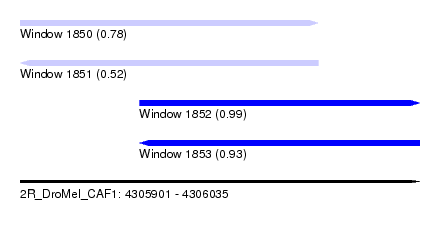
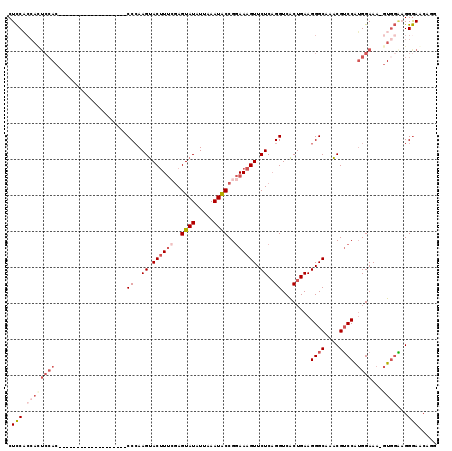
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,305,901 – 4,306,035 |
| Length | 134 |
| Max. P | 0.991877 |

| Location | 4,305,901 – 4,306,001 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 81.32 |
| Mean single sequence MFE | -32.88 |
| Consensus MFE | -21.95 |
| Energy contribution | -23.28 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.781408 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4305901 100 + 20766785 CCUGCUCCCUUCCACCUUUCCAUGGACGUUUGCCCUUCAGUGACCUGAGAACUUUCCGGUAUUUAAUAUACUCGAAAGUACUUGGG-------------------GUGGAGUGGUGGAG ..........((((((.((((((((........))........((..((.((((((.(((((.....))))).)))))).))..))-------------------)))))).)))))). ( -37.80) >DroSec_CAF1 61112 99 + 1 CCUGCUCCCUUCCAC-UUUCCAUGGACGUUUGCCCUUCAGUGACCUGAGAACUUUCCGGUAUUUAAUAUACUCGAAAGUACUUGGG-------------------GUGGAGUGGUGGAG ....((((...((((-..(((((((........))........((..((.((((((.(((((.....))))).)))))).))..))-------------------))))))))).)))) ( -34.80) >DroSim_CAF1 64508 99 + 1 CCUGCUCCCUUCCAC-UUUCCAUGGACGUUUGCCCUUCAGUGACCGGAGAACUUUCCGGUAUUUAAUAUACUCGAAAGUACUUGGG-------------------GUGGAGUGGUGGAG ....((((...((((-..(((((((........))........((.(((.((((((.(((((.....))))).)))))).))).))-------------------))))))))).)))) ( -33.60) >DroEre_CAF1 60417 91 + 1 CGUGUUCCCUUCCAC-UUUCCAUGGACGUUUGCCCUUCAGUGACCUGAGAACUUUCCGGCAUUUAAUAUACUCCAAAGUACUUGGG-------------------GU--------GGAG .(((........)))-..(((.........((((..((((....))))((....)).))))........(((((((.....)))))-------------------))--------))). ( -18.10) >DroYak_CAF1 61908 99 + 1 CAUGUUCCCUCCCAC-UUUCCAUGGACGUUUGCCCUUCAGUGACCUGAGAACUUUCCGGUAUUUAAUAUACUCGAAAGUACUUGGG-------------------GUGGAAUGGUGGAG ...........((((-(((((((((........))........((..((.((((((.(((((.....))))).)))))).))..))-------------------)))))).))))).. ( -33.30) >DroAna_CAF1 69140 115 + 1 U---UUUCCAAAUAU-UUUCCUCGGUCGCUUGCCCUUCAGCGACCUGAGAACUUUUCGGUAUUUAAUAUACUGCCAGGUGCUUUGGCGGUUAUUGGGCAGAGGGUGUGGAGUGGAGAAG (---((((((..(((-..((((((((((((........)))))))................(((((((.((((((((.....)))))))))))))))..))))).)))...))))))). ( -39.70) >consensus CCUGCUCCCUUCCAC_UUUCCAUGGACGUUUGCCCUUCAGUGACCUGAGAACUUUCCGGUAUUUAAUAUACUCGAAAGUACUUGGG___________________GUGGAGUGGUGGAG ....((((...((((...(((((((........))........((..((.((((((.(((((.....))))).)))))).))..))...................))))))))).)))) (-21.95 = -23.28 + 1.34)

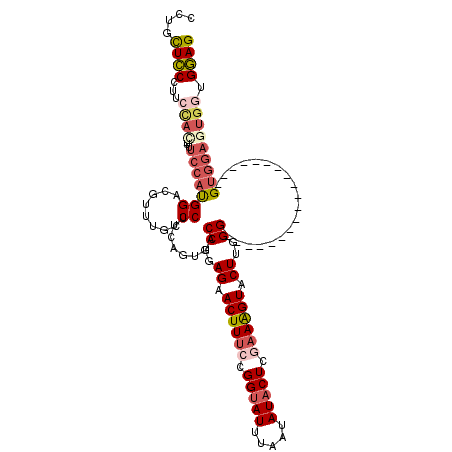
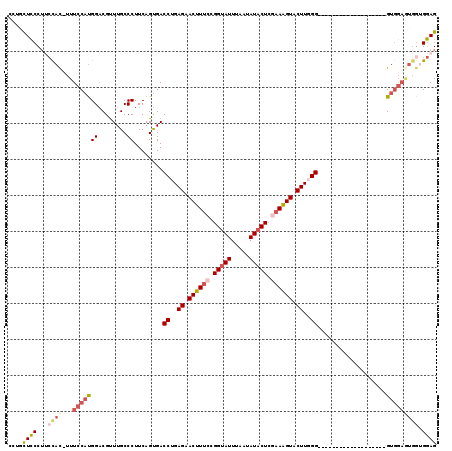
| Location | 4,305,901 – 4,306,001 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 81.32 |
| Mean single sequence MFE | -28.91 |
| Consensus MFE | -16.88 |
| Energy contribution | -19.05 |
| Covariance contribution | 2.17 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.58 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.523417 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4305901 100 - 20766785 CUCCACCACUCCAC-------------------CCCAAGUACUUUCGAGUAUAUUAAAUACCGGAAAGUUCUCAGGUCACUGAAGGGCAAACGUCCAUGGAAAGGUGGAAGGGAGCAGG ((((.....(((((-------------------((((.(.((((((..((((.....))))..)))))).)((((....)))).((((....)))).)))...))))))..)))).... ( -32.30) >DroSec_CAF1 61112 99 - 1 CUCCACCACUCCAC-------------------CCCAAGUACUUUCGAGUAUAUUAAAUACCGGAAAGUUCUCAGGUCACUGAAGGGCAAACGUCCAUGGAAA-GUGGAAGGGAGCAGG ((((.((((((((.-------------------.....(.((((((..((((.....))))..)))))).)((((....)))).((((....)))).))))..-))))...)))).... ( -30.30) >DroSim_CAF1 64508 99 - 1 CUCCACCACUCCAC-------------------CCCAAGUACUUUCGAGUAUAUUAAAUACCGGAAAGUUCUCCGGUCACUGAAGGGCAAACGUCCAUGGAAA-GUGGAAGGGAGCAGG ((((.((((((((.-------------------.....(((((....))))).......((((((......)))))).......((((....)))).))))..-))))...)))).... ( -30.00) >DroEre_CAF1 60417 91 - 1 CUCC--------AC-------------------CCCAAGUACUUUGGAGUAUAUUAAAUGCCGGAAAGUUCUCAGGUCACUGAAGGGCAAACGUCCAUGGAAA-GUGGAAGGGAACACG .(((--------..-------------------.(((((...))))).((((.....)))).)))..((((((...(((((...((((....))))......)-))))..))))))... ( -22.80) >DroYak_CAF1 61908 99 - 1 CUCCACCAUUCCAC-------------------CCCAAGUACUUUCGAGUAUAUUAAAUACCGGAAAGUUCUCAGGUCACUGAAGGGCAAACGUCCAUGGAAA-GUGGGAGGGAACAUG .(((....((((((-------------------.(((.(.((((((..((((.....))))..)))))).)((((....)))).((((....)))).)))...-)))))).)))..... ( -29.30) >DroAna_CAF1 69140 115 - 1 CUUCUCCACUCCACACCCUCUGCCCAAUAACCGCCAAAGCACCUGGCAGUAUAUUAAAUACCGAAAAGUUCUCAGGUCGCUGAAGGGCAAGCGACCGAGGAAA-AUAUUUGGAAA---A ....((((........((((.....((((((.((((.......)))).)).))))...................(((((((........)))))))))))...-.....))))..---. ( -28.79) >consensus CUCCACCACUCCAC___________________CCCAAGUACUUUCGAGUAUAUUAAAUACCGGAAAGUUCUCAGGUCACUGAAGGGCAAACGUCCAUGGAAA_GUGGAAGGGAACAGG .(((.((((((((....................((..((.((((((..((((.....))))..)))))).))..))........((((....)))).))))...))))...)))..... (-16.88 = -19.05 + 2.17)

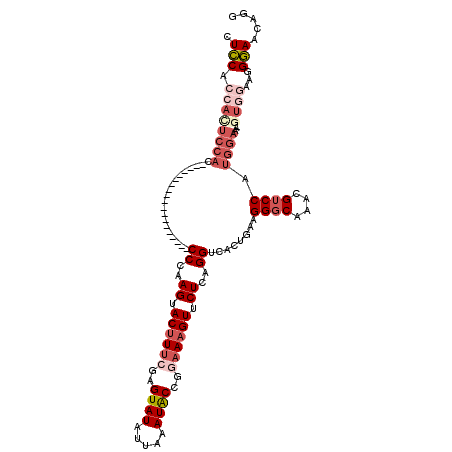
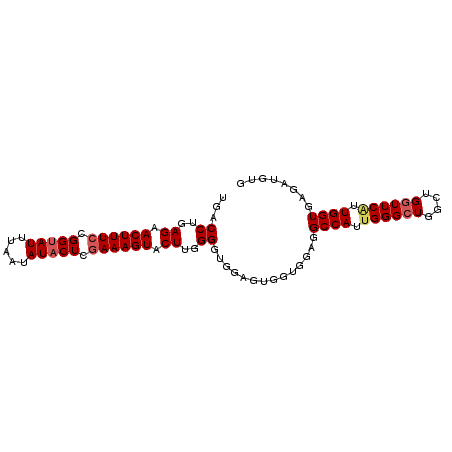
| Location | 4,305,941 – 4,306,035 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 92.66 |
| Mean single sequence MFE | -29.96 |
| Consensus MFE | -25.44 |
| Energy contribution | -26.08 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.29 |
| SVM RNA-class probability | 0.991877 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4305941 94 + 20766785 UGACCUGAGAACUUUCCGGUAUUUAAUAUACUCGAAAGUACUUGGGGUGGAGUGGUGGAGGCCAUUGGGCUGGCUGGUUCCUUGGUGAGAUGUG ...((..((.((((((.(((((.....))))).)))))).))..))..(((.((((...((((....)))).)))).))).............. ( -28.80) >DroSec_CAF1 61151 94 + 1 UGACCUGAGAACUUUCCGGUAUUUAAUAUACUCGAAAGUACUUGGGGUGGAGUGGUGGAGGCCAUUGGGCUGGCUGGUUCAUUGGUGAGAUGUG ...((..((.((((((.(((((.....))))).)))))).))..))(((((.((((...((((....)))).)))).)))))............ ( -31.50) >DroSim_CAF1 64547 94 + 1 UGACCGGAGAACUUUCCGGUAUUUAAUAUACUCGAAAGUACUUGGGGUGGAGUGGUGGAGGCCAUUGGGCUGGCUGGUUCAUUGGUGAGCUGUG ...((.(((.((((((.(((((.....))))).)))))).))).))....((((((....)))))).((((.((..(....)..)).))))... ( -32.20) >DroEre_CAF1 60456 86 + 1 UGACCUGAGAACUUUCCGGCAUUUAAUAUACUCCAAAGUACUUGGGGU--------GGAGGCCAUUGGGCUGGGUGGUUCGUUGGUGAGAUGUG (.(((...(((((.((((((........((((((((.....)))))))--------)...........)))))).)))))...))).)...... ( -28.41) >DroYak_CAF1 61947 94 + 1 UGACCUGAGAACUUUCCGGUAUUUAAUAUACUCGAAAGUACUUGGGGUGGAAUGGUGGAGGCCAUUGGGCUGGGUGUUUCGUUGGUGAGAUGUG ...((..((.((((((.(((((.....))))).)))))).))..))((..((((((....))))))..))...(..(((((....)))))..). ( -28.90) >consensus UGACCUGAGAACUUUCCGGUAUUUAAUAUACUCGAAAGUACUUGGGGUGGAGUGGUGGAGGCCAUUGGGCUGGCUGGUUCAUUGGUGAGAUGUG ...((..((.((((((.(((((.....))))).)))))).))..))..............((((.((((((....)))))).))))........ (-25.44 = -26.08 + 0.64)

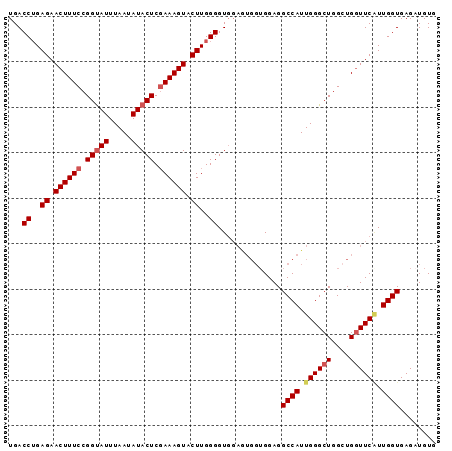
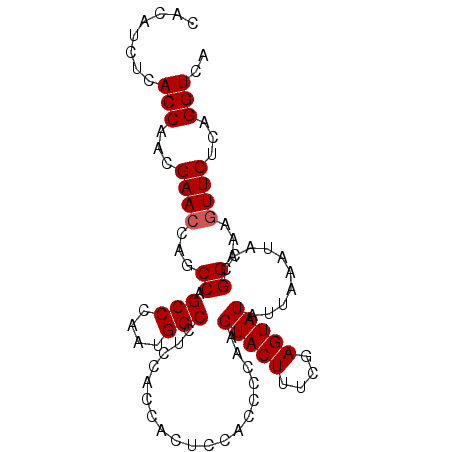
| Location | 4,305,941 – 4,306,035 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 92.66 |
| Mean single sequence MFE | -16.70 |
| Consensus MFE | -13.02 |
| Energy contribution | -13.22 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.931844 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4305941 94 - 20766785 CACAUCUCACCAAGGAACCAGCCAGCCCAAUGGCCUCCACCACUCCACCCCAAGUACUUUCGAGUAUAUUAAAUACCGGAAAGUUCUCAGGUCA .............(((....((((......)))).)))..........((..((.((((((..((((.....))))..)))))).))..))... ( -18.90) >DroSec_CAF1 61151 94 - 1 CACAUCUCACCAAUGAACCAGCCAGCCCAAUGGCCUCCACCACUCCACCCCAAGUACUUUCGAGUAUAUUAAAUACCGGAAAGUUCUCAGGUCA ........(((...((((..((((......)))).......................(((((.((((.....))))))))).))))...))).. ( -17.00) >DroSim_CAF1 64547 94 - 1 CACAGCUCACCAAUGAACCAGCCAGCCCAAUGGCCUCCACCACUCCACCCCAAGUACUUUCGAGUAUAUUAAAUACCGGAAAGUUCUCCGGUCA ....((((............((((......)))).......(((........)))......)))).........((((((......)))))).. ( -18.30) >DroEre_CAF1 60456 86 - 1 CACAUCUCACCAACGAACCACCCAGCCCAAUGGCCUCC--------ACCCCAAGUACUUUGGAGUAUAUUAAAUGCCGGAAAGUUCUCAGGUCA ........(((...((((...((.((..((((..((((--------(............)))))..))))....)).))...))))...))).. ( -15.20) >DroYak_CAF1 61947 94 - 1 CACAUCUCACCAACGAAACACCCAGCCCAAUGGCCUCCACCAUUCCACCCCAAGUACUUUCGAGUAUAUUAAAUACCGGAAAGUUCUCAGGUCA ..............((............(((((......)))))....((..((.((((((..((((.....))))..)))))).))..)))). ( -14.10) >consensus CACAUCUCACCAACGAACCAGCCAGCCCAAUGGCCUCCACCACUCCACCCCAAGUACUUUCGAGUAUAUUAAAUACCGGAAAGUUCUCAGGUCA ........(((...((((...((.(((....)))...................(((((....)))))..........))...))))...))).. (-13.02 = -13.22 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:05 2006