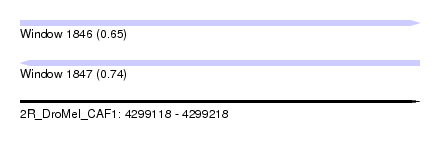
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,299,118 – 4,299,218 |
| Length | 100 |
| Max. P | 0.737665 |

| Location | 4,299,118 – 4,299,218 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 80.35 |
| Mean single sequence MFE | -38.24 |
| Consensus MFE | -23.30 |
| Energy contribution | -25.42 |
| Covariance contribution | 2.12 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.648159 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
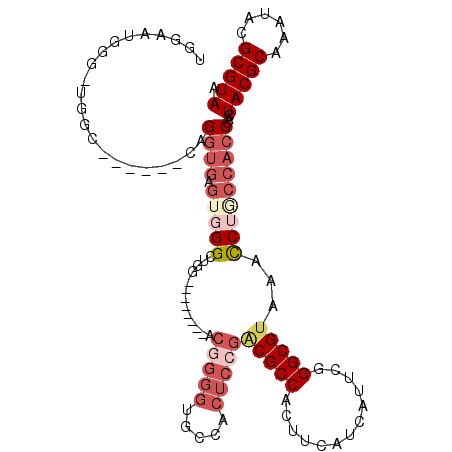
>2R_DroMel_CAF1 4299118 100 + 20766785 UUACGCGUAUUUGCGUAGUGGGUGGUUGGUUUACGCCCGAAUGAUGAAGUGGCGUCGGAGUGGCACCCAGU--------CUAGCCCACUCACCUG------GCCA-CCCAUUCCA .((((((....))))))(((((((((..(...((((((..........).)))))..(((((((.......--------...).))))))..)..------))))-))))).... ( -41.30) >DroSec_CAF1 54309 100 + 1 UUACGCGUAUUUGCGUAGUGGGUGGCAGGUUUACGCCCGAAUGAUGAAGUGGCGUCGGAGUGGCUCCCCGU--------CCAGCCCACUCACCUG------GCCA-CCCAUUCCA .((((((....))))))(((((((((((((..((((((..........).)))))..((((((((......--------..)).)))))))))).------))))-))))).... ( -45.20) >DroSim_CAF1 57735 100 + 1 UUACGCGUAUUUGCGUAGUGGGUGGCAGGUUUACGCCCGAAUGAUGAAGUGGCGUCGGAGUGGCACCCCGC--------CCAGCCCACUCACCUG------GCCA-CCCAUUCCA .((((((....))))))(((((((((((((..((((((..........).)))))..(((((((.......--------...).)))))))))).------))))-))))).... ( -43.60) >DroEre_CAF1 53837 93 + 1 UUACGCGUAUUUGCGUAGUGGGUGG--GGCUUGCGCCCGCAUGAUGAAGUGGCGUCGGAGUGGCACCCCCU--------UCAGCCC-CUCACCUG------GCCACCCCA----- .((((((....))))))(.((((((--((((.(((((.((........))))))).((((.((....))))--------))))))(-(......)------)))))))).----- ( -36.90) >DroYak_CAF1 55093 101 + 1 UUACGCGUAUUUGCGUAGUGGGUGGGAGGUUUACGCCCGAAUGAUGAAGUGGCGUCGGAGUGGCACCCCGU--------CCAGCCCACUCACCUG------CUCCACCCAUCUCC .((((((....))))))(((((((((((((..((((((..........).)))))..(((((((.......--------...).)))))))))).------).)))))))).... ( -40.00) >DroAna_CAF1 61727 101 + 1 UUACGCGUAUUUGCGUAGUGG----AAGAUUUACGCCCGAAUGAUGAAGUGGCGCC--------ACCUAGUUCCGUUAACCACCCCAUCCACCAGGAAUUUGCCA-CCCACCCC- .((((((....))))))((((----...((((......))))......((((((..--------....((((((....................)))))))))))-)))))...- ( -22.45) >consensus UUACGCGUAUUUGCGUAGUGGGUGGCAGGUUUACGCCCGAAUGAUGAAGUGGCGUCGGAGUGGCACCCCGU________CCAGCCCACUCACCUG______GCCA_CCCAUUCCA .((((((....))))))(((((((((......((((((..........).)))))..((((((.....................))))))...........)))).))))).... (-23.30 = -25.42 + 2.12)

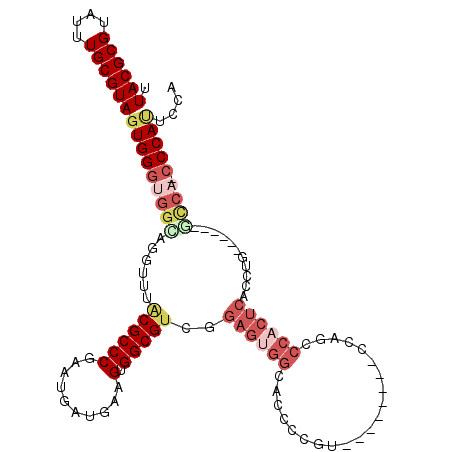
| Location | 4,299,118 – 4,299,218 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 80.35 |
| Mean single sequence MFE | -41.81 |
| Consensus MFE | -21.01 |
| Energy contribution | -23.25 |
| Covariance contribution | 2.24 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.737665 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4299118 100 - 20766785 UGGAAUGGG-UGGC------CAGGUGAGUGGGCUAG--------ACUGGGUGCCACUCCGACGCCACUUCAUCAUUCGGGCGUAAACCAACCACCCACUACGCAAAUACGCGUAA .....((((-(((.------..((((((((((((..--------....))).))))))..(((((.............)))))..)))..))))))).(((((......))))). ( -38.42) >DroSec_CAF1 54309 100 - 1 UGGAAUGGG-UGGC------CAGGUGAGUGGGCUGG--------ACGGGGAGCCACUCCGACGCCACUUCAUCAUUCGGGCGUAAACCUGCCACCCACUACGCAAAUACGCGUAA .....((((-((((------.((((((((((.((..--------......))))))))..(((((.............)))))..)))))))))))).(((((......))))). ( -43.82) >DroSim_CAF1 57735 100 - 1 UGGAAUGGG-UGGC------CAGGUGAGUGGGCUGG--------GCGGGGUGCCACUCCGACGCCACUUCAUCAUUCGGGCGUAAACCUGCCACCCACUACGCAAAUACGCGUAA .....((((-((((------.(((((((((((((..--------....))).))))))..(((((.............)))))..)))))))))))).(((((......))))). ( -44.22) >DroEre_CAF1 53837 93 - 1 -----UGGGGUGGC------CAGGUGAG-GGGCUGA--------AGGGGGUGCCACUCCGACGCCACUUCAUCAUGCGGGCGCAAGCC--CCACCCACUACGCAAAUACGCGUAA -----..(((((((------..((((((-((((.(.--------.((((......))))..)))).)))))))..))((((....)))--))))))..(((((......))))). ( -48.30) >DroYak_CAF1 55093 101 - 1 GGAGAUGGGUGGAG------CAGGUGAGUGGGCUGG--------ACGGGGUGCCACUCCGACGCCACUUCAUCAUUCGGGCGUAAACCUCCCACCCACUACGCAAAUACGCGUAA .....(((((((.(------.(((((((((((((..--------....))).))))))..(((((.............)))))..)))))))))))).(((((......))))). ( -40.62) >DroAna_CAF1 61727 101 - 1 -GGGGUGGG-UGGCAAAUUCCUGGUGGAUGGGGUGGUUAACGGAACUAGGU--------GGCGCCACUUCAUCAUUCGGGCGUAAAUCUU----CCACUACGCAAAUACGCGUAA -..((((((-.((...((.(((((..(((((((((((..((........))--------...)))))))))))..))))).))...)).)----)))))((((......)))).. ( -35.50) >consensus UGGAAUGGG_UGGC______CAGGUGAGUGGGCUGG________ACGGGGUGCCACUCCGACGCCACUUCAUCAUUCGGGCGUAAACCUGCCACCCACUACGCAAAUACGCGUAA ......................((((.(((((.............(((((.....)))))(((((.............)))))...)))))))))...(((((......))))). (-21.01 = -23.25 + 2.24)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:00 2006