| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,282,842 – 4,282,937 |
| Length | 95 |
| Max. P | 0.945769 |

| Location | 4,282,842 – 4,282,937 |
|---|---|
| Length | 95 |
| Sequences | 6 |
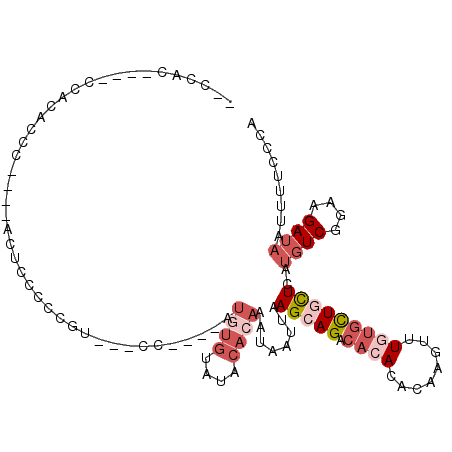
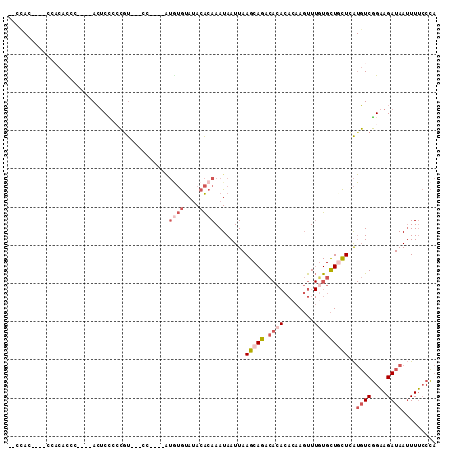
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 72.19 |
| Mean single sequence MFE | -16.68 |
| Consensus MFE | -8.23 |
| Energy contribution | -10.17 |
| Covariance contribution | 1.93 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.49 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.945769 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4282842 95 + 20766785 --CCAA----CCCCACCC----ACUCCCCCGU---CC----AUGUGUAUACACAAAUAAUUAAGCAGACACACACAAGUUUGUGCUCCUCAUGUCGGAAGAUAAUUUUCCCA --....----........----..........---..----.((((....)))).........((((((........))))))........((((....))))......... ( -13.40) >DroSec_CAF1 44177 95 + 1 --CCAC----CCCCACCC----ACUCCCCCGU---CC----AUGUGUAUACACAAAUAAUUAAGCAGACACACACAAGUUUGUGCUCCUCAUGUCGGAAGAUAAUUUUCCCA --....----........----..........---..----.((((....)))).........((((((........))))))........((((....))))......... ( -13.40) >DroEre_CAF1 43714 98 + 1 -CCCACA-CCCCACAUCC----ACUCUCCCGU---CC----AUGUGUAUACACAAAUAAUUAAGCAGACACACACAAGUUU-UGCUGCUCAUGUCGGAAGAUAAUUUUCCCA -......-..........----..(((.(((.---(.----.((((....))))........(((((....((....))..-..)))))...).))).)))........... ( -15.30) >DroYak_CAF1 44761 105 + 1 UCCCACUUUCCCACACCCU---ACUCCCCCGU---CU-AGUAUGUGUAUACACAAAUAAUUAAGCAGACACACACAAGUUUGUGCUGCUCAUGUCGGAAGAUAAUUUUCCCA ......(((((.(((....---..........---..-....((((....))))........(((((.((((........)))))))))..))).)))))............ ( -20.00) >DroMoj_CAF1 65553 83 + 1 ----------ACACACAU----AGUCGUAGGCAAACAA-------ACACACGCAAAUAAUUAAGCAGGA------AACUU-UUGUUGUUUG-CUCUGCAGAUAAAUUGACGU ----------........----.(((((((((((((((-------.((...((..........))((..------..)).-.)))))))))-).)))).))).......... ( -18.80) >DroAna_CAF1 45145 105 + 1 ACCUACCCCCCUACAUGUUGCAACACCCACAU---AU----GUAUGUAUACACAAAUAAUUAAGCAGACACACACAAGUUUGUGCUGCUCAUGUCGGAAGAUAAUUUUCCCA .........((.(((((.(((((((.......---.)----)).))))..............(((((.((((........)))))))))))))).))............... ( -19.20) >consensus __CCAC____CCACACCC____ACUCCCCCGU___CC____AUGUGUAUACACAAAUAAUUAAGCAGACACACACAAGUUUGUGCUGCUCAUGUCGGAAGAUAAUUUUCCCA ..........................................((((....))))........(((((.((((........)))))))))..((((....))))......... ( -8.23 = -10.17 + 1.93)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:54:57 2006