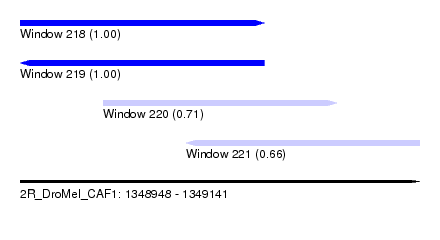
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,348,948 – 1,349,141 |
| Length | 193 |
| Max. P | 0.999568 |

| Location | 1,348,948 – 1,349,066 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.13 |
| Mean single sequence MFE | -45.05 |
| Consensus MFE | -39.98 |
| Energy contribution | -40.78 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.97 |
| SVM RNA-class probability | 0.997960 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1348948 118 + 20766785 UGGGCGUGUCUGCGGGGUGUCUGCACAUAUUUAUGAGCAGCGAUGUGCUGCGACGUCGUCCGACGUUGAAUUAAUGAAAUACCUGCCAAGAUUUAUGUGCCAGCUGACUGGCACA--GGG .......((((((((((((....)))...(((((..(((((.....)))))((((((....))))))......)))))...)))))..))))...((((((((....))))))))--... ( -42.40) >DroSec_CAF1 67207 120 + 1 UGGGCGUGUCUGCGGGGUGUCUGCACAUAUUUAUGAGCAGCGAUGCGCUGCGACGUCGUCCGACGUUGAAUUAAUGAAAUACCUGCCAAGAUUUAUGUGCCAGCUGACUGGCACACGGGG .(((((((((.((((.((((((((.(((....))).)))..))))).)))))))).)))))..(((((...))))).....((((..........((((((((....)))))))))))). ( -45.30) >DroSim_CAF1 45342 120 + 1 UGGGCGUGUCUGCGGGGUGUCUGCACAUAUUUAUGAGCAGCGAUGCGCUGCGACGUCGUCCGACGUUGAAUUAAUGAAAUACCUGCCAAGAUUUAUGUGCCAGCUGACUGGCACACGGGG .(((((((((.((((.((((((((.(((....))).)))..))))).)))))))).)))))..(((((...))))).....((((..........((((((((....)))))))))))). ( -45.30) >DroEre_CAF1 47821 118 + 1 UGGGCGUGUCUGCGGGGUGUCUGCACAUAUUUAUGAGCAGUGAUGCGCUGCGACGUCGUCCGACGUGGAAUUAAUGAAAUACCUGCCACGAUUUAUGUGCCAGGUGACUGGCACA--GGG .(((((((((.((((.((((((((.(((....))).)))..))))).)))))))).)))))..(((((.................))))).....((((((((....))))))))--... ( -52.73) >DroYak_CAF1 35293 111 + 1 -------GUCUGCGGGGUGUCUGCACAUAUUUAUGAGCAGUGAUGCGGUGCGACGUCGCUUGACGUUGAAAUAAUGAAAUACCUGCCAAGAUUUAUGUGCCAGCUGACUGGCACA--GGG -------((((((((((((....))).(((((....(((....)))....(((((((....))))))))))))........)))))..))))...((((((((....))))))))--... ( -39.50) >consensus UGGGCGUGUCUGCGGGGUGUCUGCACAUAUUUAUGAGCAGCGAUGCGCUGCGACGUCGUCCGACGUUGAAUUAAUGAAAUACCUGCCAAGAUUUAUGUGCCAGCUGACUGGCACA__GGG .......((((((((((((....)))...(((((..((((((...))))))((((((....))))))......)))))...)))))..))))...((((((((....))))))))..... (-39.98 = -40.78 + 0.80)

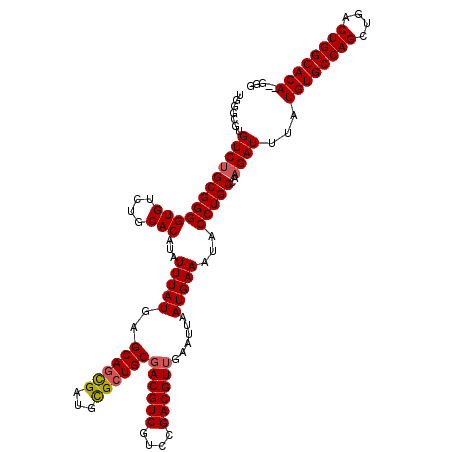
| Location | 1,348,948 – 1,349,066 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.13 |
| Mean single sequence MFE | -37.10 |
| Consensus MFE | -33.26 |
| Energy contribution | -33.86 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.73 |
| SVM RNA-class probability | 0.999568 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1348948 118 - 20766785 CCC--UGUGCCAGUCAGCUGGCACAUAAAUCUUGGCAGGUAUUUCAUUAAUUCAACGUCGGACGACGUCGCAGCACAUCGCUGCUCAUAAAUAUGUGCAGACACCCCGCAGACACGCCCA ...--((((((((....)))))))).........(((.((((((..........(((((....))))).(((((.....)))))....)))))).))).........((......))... ( -37.00) >DroSec_CAF1 67207 120 - 1 CCCCGUGUGCCAGUCAGCUGGCACAUAAAUCUUGGCAGGUAUUUCAUUAAUUCAACGUCGGACGACGUCGCAGCGCAUCGCUGCUCAUAAAUAUGUGCAGACACCCCGCAGACACGCCCA ....(((((((((....)))))))))........(((.((((((..........(((((....))))).((((((...))))))....)))))).))).........((......))... ( -39.50) >DroSim_CAF1 45342 120 - 1 CCCCGUGUGCCAGUCAGCUGGCACAUAAAUCUUGGCAGGUAUUUCAUUAAUUCAACGUCGGACGACGUCGCAGCGCAUCGCUGCUCAUAAAUAUGUGCAGACACCCCGCAGACACGCCCA ....(((((((((....)))))))))........(((.((((((..........(((((....))))).((((((...))))))....)))))).))).........((......))... ( -39.50) >DroEre_CAF1 47821 118 - 1 CCC--UGUGCCAGUCACCUGGCACAUAAAUCGUGGCAGGUAUUUCAUUAAUUCCACGUCGGACGACGUCGCAGCGCAUCACUGCUCAUAAAUAUGUGCAGACACCCCGCAGACACGCCCA ...--((((((((....)))))))).....((((((.((.(((.....))).))(((((....))))).)).(((.....((((.(((....))).))))......)))...)))).... ( -37.20) >DroYak_CAF1 35293 111 - 1 CCC--UGUGCCAGUCAGCUGGCACAUAAAUCUUGGCAGGUAUUUCAUUAUUUCAACGUCAAGCGACGUCGCACCGCAUCACUGCUCAUAAAUAUGUGCAGACACCCCGCAGAC------- ...--((((((((....))))))))....((((((..(((..............(((((....)))))............((((.(((....))).))))..)))))).))).------- ( -32.30) >consensus CCC__UGUGCCAGUCAGCUGGCACAUAAAUCUUGGCAGGUAUUUCAUUAAUUCAACGUCGGACGACGUCGCAGCGCAUCGCUGCUCAUAAAUAUGUGCAGACACCCCGCAGACACGCCCA .....((((((((....)))))))).........(((.((((((..........(((((....))))).(((((.....)))))....)))))).)))...................... (-33.26 = -33.86 + 0.60)

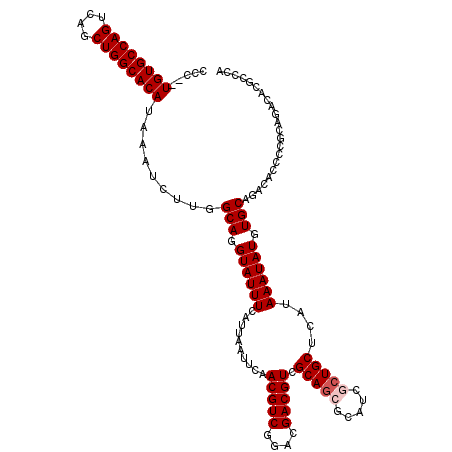
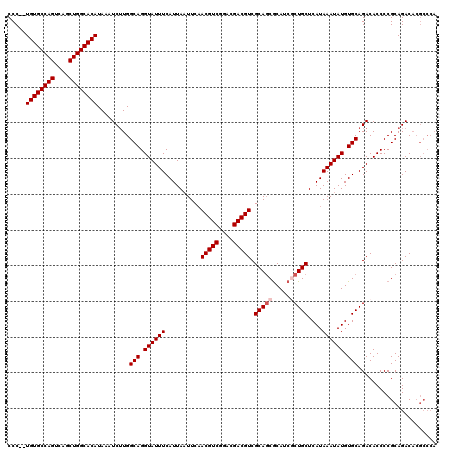
| Location | 1,348,988 – 1,349,101 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.52 |
| Mean single sequence MFE | -41.88 |
| Consensus MFE | -29.74 |
| Energy contribution | -31.34 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.714407 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1348988 113 + 20766785 CGAUGUGCUGCGACGUCGUCCGACGUUGAAUUAAUGAAAUACCUGCCAAGAUUUAUGUGCCAGCUGACUGGCACA--GGGGAC-----ACACGCGUGAACGGGCGAAGGCGGGAAAAGAA ..........(((((((....))))))).............((((((........((((((((....))))))))--(....)-----...(((.(....).)))..))))))....... ( -40.70) >DroSec_CAF1 67247 115 + 1 CGAUGCGCUGCGACGUCGUCCGACGUUGAAUUAAUGAAAUACCUGCCAAGAUUUAUGUGCCAGCUGACUGGCACACGGGGGAC-----ACACGCGUGAACGGGCGGGGGAGGGAAAAGAA .....(.((.(((((((....))))))).............((((((....((..((((((((....))))))))..))(..(-----((....)))..).))))))..)).)....... ( -39.70) >DroSim_CAF1 45382 115 + 1 CGAUGCGCUGCGACGUCGUCCGACGUUGAAUUAAUGAAAUACCUGCCAAGAUUUAUGUGCCAGCUGACUGGCACACGGGGGAC-----ACACGCGUGAACGGGCGGGGGAGGGAAAAGAA .....(.((.(((((((....))))))).............((((((....((..((((((((....))))))))..))(..(-----((....)))..).))))))..)).)....... ( -39.70) >DroEre_CAF1 47861 118 + 1 UGAUGCGCUGCGACGUCGUCCGACGUGGAAUUAAUGAAAUACCUGCCACGAUUUAUGUGCCAGGUGACUGGCACA--GGGGACACCCGACACACGUGGUCGGGCGGAGGAGCGAAAAGAA .....((((.(....(((((((((..((.(((.....))).))...((((.....((((((((....))))))))--(((....)))......))))))))))))).).))))....... ( -52.30) >DroYak_CAF1 35326 111 + 1 UGAUGCGGUGCGACGUCGCUUGACGUUGAAAUAAUGAAAUACCUGCCAAGAUUUAUGUGCCAGCUGACUGGCACA--GGGGAC-------ACACGUGAACUGGCGAGGGAGCGAAAAGAA ...(((....(((((((....))))))).............(((((((.(.((((((((((((....))))))).--(....)-------....)))))))))).)))..)))....... ( -37.00) >consensus CGAUGCGCUGCGACGUCGUCCGACGUUGAAUUAAUGAAAUACCUGCCAAGAUUUAUGUGCCAGCUGACUGGCACA__GGGGAC_____ACACGCGUGAACGGGCGGGGGAGGGAAAAGAA ..........(((((((....))))))).............((((((........((((((((....))))))))..(....)..................))))))............. (-29.74 = -31.34 + 1.60)

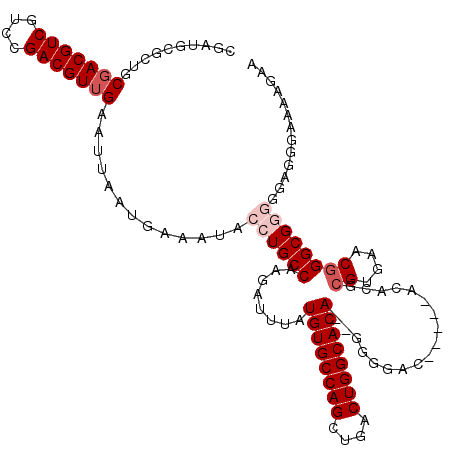
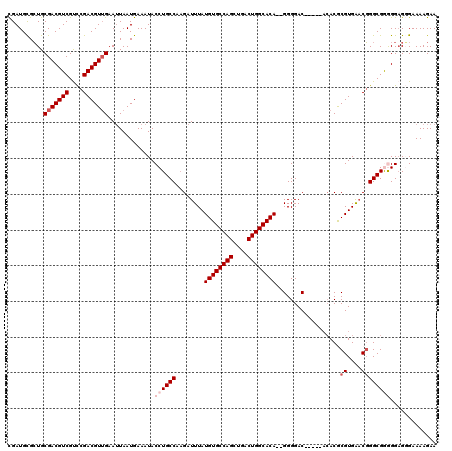
| Location | 1,349,028 – 1,349,141 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.18 |
| Mean single sequence MFE | -28.78 |
| Consensus MFE | -20.30 |
| Energy contribution | -20.86 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.661524 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1349028 113 - 20766785 CUAAUAAAACGAUUCAGCACUAUUGCCCUGCAAAAUUAAUUUCUUUUCCCGCCUUCGCCCGUUCACGCGUGU-----GUCCCC--UGUGCCAGUCAGCUGGCACAUAAAUCUUGGCAGGU ................(((....)))(((((..................(((...(((........))).))-----).....--((((((((....)))))))).........))))). ( -28.00) >DroSec_CAF1 67287 115 - 1 CUAAUAAAACGAUUCAGCACUAUUGCCCUGCAAAAUUAAUUUCUUUUCCCUCCCCCGCCCGUUCACGCGUGU-----GUCCCCCGUGUGCCAGUCAGCUGGCACAUAAAUCUUGGCAGGU ................(((....)))(((((........................(((.(((....))).))-----)......(((((((((....)))))))))........))))). ( -29.50) >DroSim_CAF1 45422 115 - 1 CUAAUAAAACGAUUCAGCACUAUUGCCCUGCAAAAUUAAUUUCUUUUCCCUCCCCCGCCCGUUCACGCGUGU-----GUCCCCCGUGUGCCAGUCAGCUGGCACAUAAAUCUUGGCAGGU ................(((....)))(((((........................(((.(((....))).))-----)......(((((((((....)))))))))........))))). ( -29.50) >DroEre_CAF1 47901 118 - 1 CUAAUACGACGUUUCACCACUAUUCCCCCACAAAAUUAAUUUCUUUUCGCUCCUCCGCCCGACCACGUGUGUCGGGUGUCCCC--UGUGCCAGUCACCUGGCACAUAAAUCGUGGCAGGU ........................((.((((........................(((((((((....).)))))))).....--((((((((....))))))))......))))..)). ( -33.80) >DroYak_CAF1 35366 111 - 1 CUCAUAAAACGAUUCAGCACUAUUCCCCCACAAAAUUAAUUUCUUUUCGCUCCCUCGCCAGUUCACGUGU-------GUCCCC--UGUGCCAGUCAGCUGGCACAUAAAUCUUGGCAGGU ....................................................(((.(((((..((....)-------)..)..--((((((((....)))))))).......))))))). ( -23.10) >consensus CUAAUAAAACGAUUCAGCACUAUUGCCCUGCAAAAUUAAUUUCUUUUCCCUCCCCCGCCCGUUCACGCGUGU_____GUCCCC__UGUGCCAGUCAGCUGGCACAUAAAUCUUGGCAGGU ..........................(((((........................(((........)))................((((((((....)))))))).........))))). (-20.30 = -20.86 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:27:19 2006