| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,241,516 – 4,241,634 |
| Length | 118 |
| Max. P | 0.559410 |

| Location | 4,241,516 – 4,241,634 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.03 |
| Mean single sequence MFE | -32.22 |
| Consensus MFE | -18.62 |
| Energy contribution | -18.77 |
| Covariance contribution | 0.15 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.559410 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
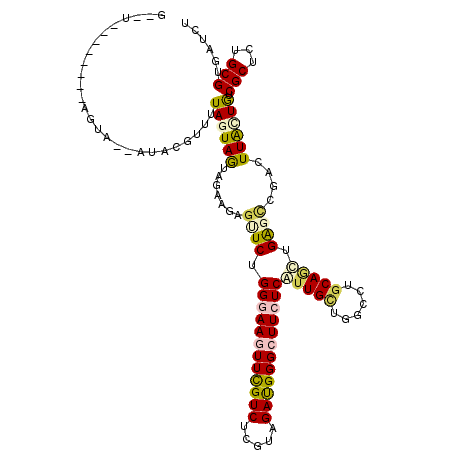
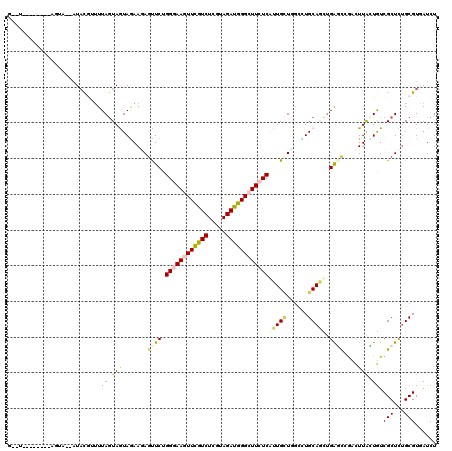
>2R_DroMel_CAF1 4241516 118 + 20766785 GUUUUCGGUUCGAGGG--AUACGGUUUAGUAGUAGAAGAGUUCCGGAAAGUUCGUCUCGUAGAUGGGCUUCUCAUUGCUGGCUUGCAACUGACUGGACUUGGUGUCGCUUUGCGUGAUCU ......((((((((((--(((((((((((((((...........(..(((((((((.....)))))))))..)..(((......)))))).))))))))..))))).)))))...)))). ( -34.20) >DroVir_CAF1 5609 105 + 1 ---------------AAUAUAUGUUUUAGUAGUAGAACAGCUCUGGGAAUUUCGUCUCGUAGACGGGUUUCUCAUUGUUGGUCUGCAGUGGGGCUGUCUUAUUGUCGCUCAGCGUAAUCU ---------------....((((((...((((((..(((((((((((((.((((((.....)))))).)))))(((((......)))))))))))))..))))).)....)))))).... ( -33.40) >DroYak_CAF1 3027 110 + 1 --------UUCGAGGA--AUACGUUUUAAUAGUAAAAGAGUUCCGGGAAGUUCGUCUCGUAGAUGGGCUUCUCAUUGCUAGCUUGCAACUGACUGGACUUAGUGUCGCUCUGCGUGAUCU --------...(((..--((((.(((((....)))))((((.((((((((((((((.....))))))))))(((((((......)))).))))))))))).))))..))).......... ( -31.80) >DroMoj_CAF1 2785 113 + 1 UUUUA-------UAUAAUUUUCGUUUUAGUAGUAGAAGAGCUCUGGGAAUUUCGUCUCAUAGACGGGCUUCUCAUUGGUUGUCUGCAUUUGAGCUGUUUUGCUAUCGCUUUGCGUUAUCU .....-------.........(((....(((((((((.(((((((((((.((((((.....)))))).))))))...((.....))....))))).)))))))))......)))...... ( -31.50) >DroAna_CAF1 3055 116 + 1 G--UCCGGUUUACAUA--AUACCAUUUAGUAAUAAAAGAGUUCGGGGAAGUUCGUCUCAUAGAUGGGCUUCUCAUUGCUCGCCUGCAGUUGAGCCGCCUUAUUGUCGCUCUGCGUGAUCU (--((((((((((.((--(......))))))).....((((...((((((((((((.....))))))))))))...))))))).(((...((((.((......)).)))))))).))).. ( -34.80) >DroPer_CAF1 7275 112 + 1 GUUCAC------AGUA--AUACAGAUUAGUAAUAGAAUAAUUCUGGAAAGUUUGUCUCGUAGAUGGGCUUCUCGUUGCUGCCCUGCAGCUGGGCCACCUUGCUGUCGCUUUGCGUUAUCU ....((------((((--(........(((((((((.....)))(..(((((..((.....))..)))))..)))))))((((.......))))....)))))))(((...)))...... ( -27.60) >consensus G__U________AGUA__AUACGUUUUAGUAGUAGAAGAGUUCUGGGAAGUUCGUCUCGUAGAUGGGCUUCUCAUUGCUGGCCUGCAGCUGAGCCGACUUACUGUCGCUCUGCGUGAUCU ..........................((((((.......((((.((((((((((((.....))))))))))))(((((......))))).))))....)))))).(((...)))...... (-18.62 = -18.77 + 0.15)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:54:47 2006