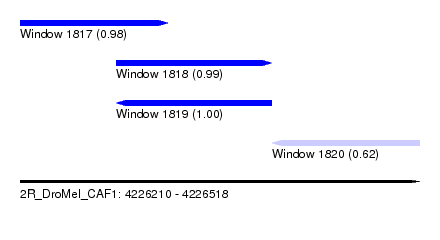
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,226,210 – 4,226,518 |
| Length | 308 |
| Max. P | 0.996881 |

| Location | 4,226,210 – 4,226,324 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.50 |
| Mean single sequence MFE | -27.46 |
| Consensus MFE | -23.36 |
| Energy contribution | -23.76 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.979837 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4226210 114 + 20766785 GACUGGGAUAUU------AAAUACCGCCAAAACCAGCGGUCAACUUCACAAUCGGUAGCAGACCAGAGCCAACUAUUUGCCACUUGAACCGCAGUCUCAUUAACGACAUCCUCGUUAACA (((((((.....------....(((((........)))))...........(((((.(((((..((......)).))))).)).))).)).)))))...(((((((.....))))))).. ( -27.10) >DroSec_CAF1 38077 114 + 1 GACUGGGAUAUU------AAAUACCGCCAAAAGCAGCGGUCAACUUCACAAUCGGUCGCAGACCAGAGCCAACUAUUUGCCACUUGAACCGCAGUCUCAUUAACGACAUCCUCGUUAACA (((((((.....------....(((((........)))))...........(((((.(((((..((......)).))))).)).))).)).)))))...(((((((.....))))))).. ( -27.10) >DroSim_CAF1 67479 114 + 1 GACUGGGAUAUU------AAAUACCGCCAAAAGCAGCGGUCAACUUCACAAUCGGUCGCAGACCAGAGCCAACUAUUUGCCACUUGAACCGCAGUCUCAUUAACGACAUCCUCGUUAACA (((((((.....------....(((((........)))))...........(((((.(((((..((......)).))))).)).))).)).)))))...(((((((.....))))))).. ( -27.10) >DroEre_CAF1 39588 114 + 1 GACUGAGAUAUU------AAAUACCGCCAAAAGCAGCGGUCAACUUCACAAUCGGUCGCAGAGCAGAGCAAACUAUUUGCCACUUGAACCGCAGUCUCAUUAACGACAUCCUCGUUAACA (((((.......------....(((((........)))))............((((((.((.(((((........)))))..)))).)))))))))...(((((((.....))))))).. ( -27.00) >DroYak_CAF1 40721 120 + 1 CACUGAGAUAUUACUCGUAAAUACCGCCAAAAGCAGCGGUCAGCUUUACAAUCGGUCACAGACCAGAGCAAACUAUUUGCCACUUGAACCGCAGUCUCAUUAACGACAUCCUCGUUAACA ...((((((................((.....)).((((((((..........((((...)))).(.((((.....)))))..))).))))).))))))(((((((.....))))))).. ( -29.00) >consensus GACUGGGAUAUU______AAAUACCGCCAAAAGCAGCGGUCAACUUCACAAUCGGUCGCAGACCAGAGCCAACUAUUUGCCACUUGAACCGCAGUCUCAUUAACGACAUCCUCGUUAACA (((((.(...............(((((........)))))...........(((((.(((((..((......)).))))).)).)))..).)))))...(((((((.....))))))).. (-23.36 = -23.76 + 0.40)

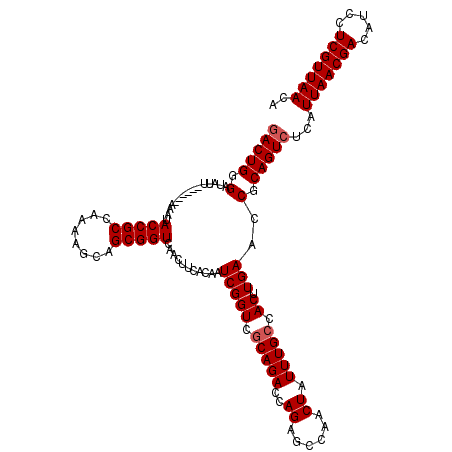
| Location | 4,226,284 – 4,226,404 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.15 |
| Mean single sequence MFE | -24.86 |
| Consensus MFE | -24.60 |
| Energy contribution | -24.60 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991216 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4226284 120 + 20766785 CACUUGAACCGCAGUCUCAUUAACGACAUCCUCGUUAACAAAGUUGCUCUGAAUGGGUUUCAGUUGUUUUUGUGCUUAAUAACUUGCUUAAGCUUUCAGUUUUUAAGUUCAACUCCGUCC ...((((((.((((..(..(((((((.....)))))))..)..)))).(((((..(((((.(((.(((.(((....))).)))..))).)))))))))).......))))))........ ( -25.40) >DroSec_CAF1 38151 120 + 1 CACUUGAACCGCAGUCUCAUUAACGACAUCCUCGUUAACAAAGUUGCUCUGAAUGGGUUUCAGUUGUUUUUGUGCUUAAUAACUUGCUUAAGCUUUCAGUUUUUAAGUUCACCUCCGUCC (((..((((.((((..(..(((((((.....)))))))..)..)))).(((((.....)))))..))))..)))(((((.((((.((....))....)))).)))))............. ( -24.60) >DroSim_CAF1 67553 120 + 1 CACUUGAACCGCAGUCUCAUUAACGACAUCCUCGUUAACAAAGUUGCUCUGAAUGGGUUUCAGUUGUUUUUGUGCUUAAUAACUUGCUUAAGCUUUCAGUUUUUAAGUUCACCUCCGUCC (((..((((.((((..(..(((((((.....)))))))..)..)))).(((((.....)))))..))))..)))(((((.((((.((....))....)))).)))))............. ( -24.60) >DroEre_CAF1 39662 116 + 1 CACUUGAACCGCAGUCUCAUUAACGACAUCCUCGUUAACAAAGUUGCUCUGAAUGAGUUUCAGUUGUUUUUGUGCUUAAUAACUUGCUUAAGCUUUCAGUUUUUAAGUUCACU----CCC ....(((((.((((..(..(((((((.....)))))))..)..)))).((((..((((((.(((.(((.(((....))).)))..))).)))))))))).......)))))..----... ( -25.00) >DroYak_CAF1 40801 116 + 1 CACUUGAACCGCAGUCUCAUUAACGACAUCCUCGUUAACAAAGUUGCUCUGAAUGAGUUUCAGUUGUUUUUGUGCUUAAUAACUUGCUUAAGCUUUCAGUUUUUAAGUUCCCC----UCC .(((((((..((((..(..(((((((.....)))))))..)..)))).((((..((((((.(((.(((.(((....))).)))..))).))))))))))..))))))).....----... ( -24.70) >consensus CACUUGAACCGCAGUCUCAUUAACGACAUCCUCGUUAACAAAGUUGCUCUGAAUGGGUUUCAGUUGUUUUUGUGCUUAAUAACUUGCUUAAGCUUUCAGUUUUUAAGUUCACCUCCGUCC (((..((((.((((..(..(((((((.....)))))))..)..)))).(((((.....)))))..))))..)))(((((.((((.((....))....)))).)))))............. (-24.60 = -24.60 + -0.00)

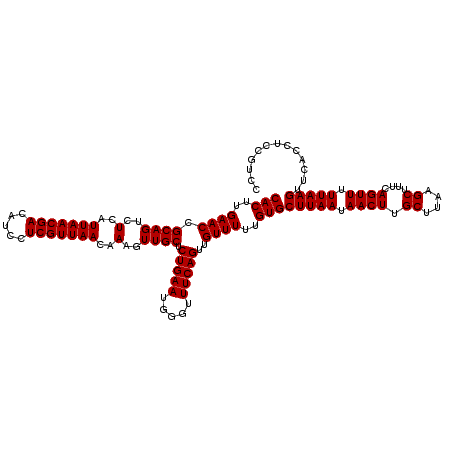
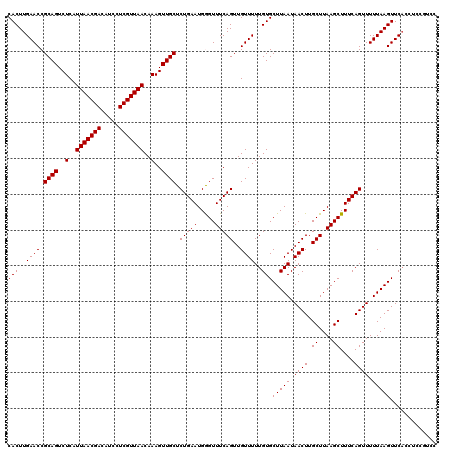
| Location | 4,226,284 – 4,226,404 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.15 |
| Mean single sequence MFE | -28.28 |
| Consensus MFE | -27.08 |
| Energy contribution | -27.28 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.76 |
| SVM RNA-class probability | 0.996881 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4226284 120 - 20766785 GGACGGAGUUGAACUUAAAAACUGAAAGCUUAAGCAAGUUAUUAAGCACAAAAACAACUGAAACCCAUUCAGAGCAACUUUGUUAACGAGGAUGUCGUUAAUGAGACUGCGGUUCAAGUG ......(.(((((((......(((((.((((((........)))))).((........)).......))))).(((..((..(((((((.....)))))))..))..)))))))))).). ( -29.30) >DroSec_CAF1 38151 120 - 1 GGACGGAGGUGAACUUAAAAACUGAAAGCUUAAGCAAGUUAUUAAGCACAAAAACAACUGAAACCCAUUCAGAGCAACUUUGUUAACGAGGAUGUCGUUAAUGAGACUGCGGUUCAAGUG ((((....(((..(((((.((((....((....)).)))).))))))))........(((((.....))))).(((..((..(((((((.....)))))))..))..))).))))..... ( -28.40) >DroSim_CAF1 67553 120 - 1 GGACGGAGGUGAACUUAAAAACUGAAAGCUUAAGCAAGUUAUUAAGCACAAAAACAACUGAAACCCAUUCAGAGCAACUUUGUUAACGAGGAUGUCGUUAAUGAGACUGCGGUUCAAGUG ((((....(((..(((((.((((....((....)).)))).))))))))........(((((.....))))).(((..((..(((((((.....)))))))..))..))).))))..... ( -28.40) >DroEre_CAF1 39662 116 - 1 GGG----AGUGAACUUAAAAACUGAAAGCUUAAGCAAGUUAUUAAGCACAAAAACAACUGAAACUCAUUCAGAGCAACUUUGUUAACGAGGAUGUCGUUAAUGAGACUGCGGUUCAAGUG ...----..((((((......(((((.((((((........))))))...........((.....))))))).(((..((..(((((((.....)))))))..))..))))))))).... ( -28.10) >DroYak_CAF1 40801 116 - 1 GGA----GGGGAACUUAAAAACUGAAAGCUUAAGCAAGUUAUUAAGCACAAAAACAACUGAAACUCAUUCAGAGCAACUUUGUUAACGAGGAUGUCGUUAAUGAGACUGCGGUUCAAGUG ...----...(((((......(((((.((((((........))))))...........((.....))))))).(((..((..(((((((.....)))))))..))..))))))))..... ( -27.20) >consensus GGACGGAGGUGAACUUAAAAACUGAAAGCUUAAGCAAGUUAUUAAGCACAAAAACAACUGAAACCCAUUCAGAGCAACUUUGUUAACGAGGAUGUCGUUAAUGAGACUGCGGUUCAAGUG .........((((((......(((((.((((((........)))))).((........)).......))))).(((..((..(((((((.....)))))))..))..))))))))).... (-27.08 = -27.28 + 0.20)

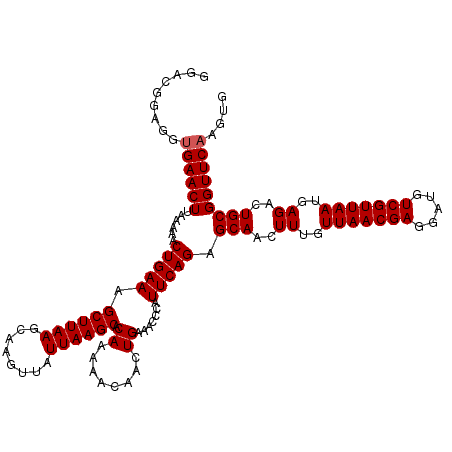
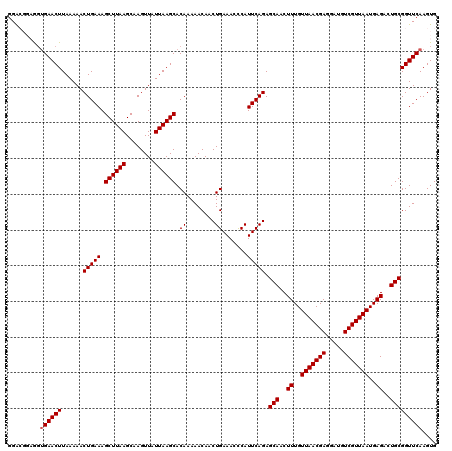
| Location | 4,226,404 – 4,226,518 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.50 |
| Mean single sequence MFE | -38.39 |
| Consensus MFE | -35.26 |
| Energy contribution | -35.74 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.615066 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4226404 114 - 20766785 ------UGCAGCUCGAAAGACCUGCAGAGGGACAGCAAGGUGAGUGCGUUGGGCUGUAACCACUUUGUCAAUCCACUUUGUCCAACCAACCGAGUGCCACCAAGUGCUUUCCUUAUGGCA ------(((((.((....)).))))).(((((.((((.((((.(..((((((((.........................)))))))(....).)..)))))...)))).)))))...... ( -38.21) >DroSec_CAF1 38271 120 - 1 AUAAUCUGCAGCUCGAAGGACCUGCAGAGGGACAGCAAGGUGAGUGCGUUGGGCUGGAACCACUUUGUCAAUCCACUUUGUCCAACCAACCGAGUGCCACCAAGUGCUUUCCUUGUGGCA ....(((((((.((....)).)))))))((((.((((.((((.(..((((((((((((.............))))....)))))))(....).)..)))))...)))).))))....... ( -43.32) >DroSim_CAF1 67673 120 - 1 AUAAUCUGCAGCUCGAAGGACCUGCAGAGGGACAGCAAGGUGAGUGCGUUGGGCUGGAACCACUUUGUCAAUCCACUUUGUCCAACCAACCGAGUGUUACCAAGUGCUUUCCUUGUGGCA ....(((((((.((....)).)))))))..(...((((((.(((..((((((((((((.............))))....))))))).(((.....))).....)..))).))))))..). ( -40.52) >DroEre_CAF1 39778 119 - 1 AUCAUCUGAAGCUCGAAGGACCUGCAGAAGGAUAGCA-GGUGAGUGCGUUGGGCUGGAACCACUUUGUCAAUCUACUUUGUCCAACCAACCGAGUGCCACCAAGUGCUUUCCUUGUGGCA .(((((((.((.((....)).)).)))(((((.((((-((((.(..((((((..((((........(.....).......)))).)))))...)..)))))...)))).))))))))).. ( -35.66) >DroYak_CAF1 40917 120 - 1 AUAAUCUGAAGCUCGAAGGACCUGCAGAAGGAUAGCAAGGUGAGUGCGUUGGGCUGGAACCACUUUGUCAAUCUACUUUGUCCAACCAACCGAGUGGCACCAAGUGCUUUCCUUGUGGCA ....((((.((.((....)).)).))))((((.((((.((((..((.(((((..((((........(.....).......)))).))))))).....))))...)))).))))....... ( -34.26) >consensus AUAAUCUGCAGCUCGAAGGACCUGCAGAGGGACAGCAAGGUGAGUGCGUUGGGCUGGAACCACUUUGUCAAUCCACUUUGUCCAACCAACCGAGUGCCACCAAGUGCUUUCCUUGUGGCA .....((((((.((....)).))))))(((((.((((.((((.(((((((((..((((..........(((......))))))).)))))...))))))))...)))).)))))...... (-35.26 = -35.74 + 0.48)

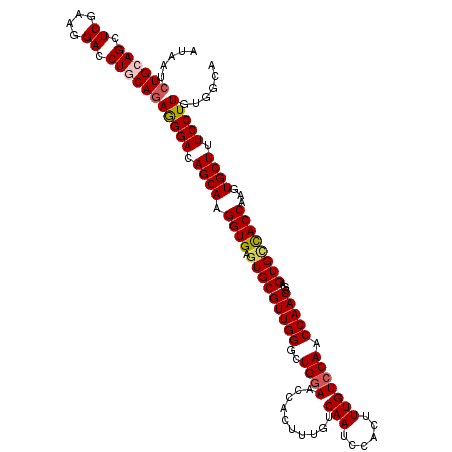
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:54:33 2006