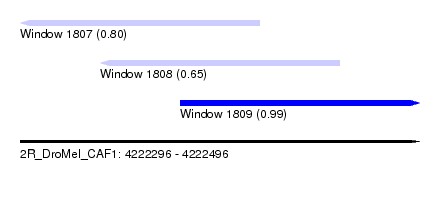
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,222,296 – 4,222,496 |
| Length | 200 |
| Max. P | 0.988029 |

| Location | 4,222,296 – 4,222,416 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -33.46 |
| Consensus MFE | -27.80 |
| Energy contribution | -29.16 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.798493 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4222296 120 - 20766785 AUGACGGUGUAACCAUCUACAUAUUCCUGGUGAUAACCAUCACAACCACCUUGUGGUGGUGCCUCACCUUCAACUUUGUUGCCCGCCGAUCCGCUGGGCUGUUCAUGUGGUCCCUAUUCG ....(((.(..(((((..(((.......(((((..((((((((((.....))))))))))...)))))............((((((......)).)))))))....)))))..)...))) ( -36.90) >DroSec_CAF1 34012 120 - 1 AUGACGGUGUAACCAUCUACAUAUUCCUGGUGAUAACCAUCACAACCACCUUGUGGUGGAGCCUCACCUUCAACUUUGUUGCCCGCCGAUCCGCUGGGCUGUUCAUGUGGUCCCUAUUCG ....(((.(..(((((..(((.......(((((...(((((((((.....)))))))))....)))))............((((((......)).)))))))....)))))..)...))) ( -33.90) >DroSim_CAF1 61733 120 - 1 AUGACGGUGUAACCAUCUACAUAUUCCUGGUGAUAACCAUCACAACCACCUUGUGGUGGAGCCUCACCUUCAACUUUGUUGCCCGCCGAUCCGCUGGGCUGUUCAUGUGGUCCCUAUUCG ....(((.(..(((((..(((.......(((((...(((((((((.....)))))))))....)))))............((((((......)).)))))))....)))))..)...))) ( -33.90) >DroEre_CAF1 35319 120 - 1 AUGAAGGCGUAACCAUCUACAUAUUCCUCGUGAUAACCAUCACCACCACCUUGUGGUGGUGCCUCACCUUCAACUUUGUUGCCCGCCGAUCCGCUGGCUUGUUCAUGUGGUCCAUAUUCA .((((((((((((................(((((....)))))(((((((....)))))))................)))))..)))((.(((((((.....))).))))))....)))) ( -31.30) >DroYak_CAF1 36549 120 - 1 ACGAAGGCGUUACCAUCUACAUAUUCCUGAUAAUAACCAUCACAACCACCUUGUGGUGGUGCCUCACAUUCAACUUCGUUGCCCGCCGAUCCGCUGGGUUGUUCAUGUGGAGCAUAUUCA ((((((..((........)).......(((.....((((((((((.....))))))))))...))).......))))))(((((((((((((...)))))).....)))).)))...... ( -31.30) >consensus AUGACGGUGUAACCAUCUACAUAUUCCUGGUGAUAACCAUCACAACCACCUUGUGGUGGUGCCUCACCUUCAACUUUGUUGCCCGCCGAUCCGCUGGGCUGUUCAUGUGGUCCCUAUUCG .....((.....))..((((((......(((((..((((((((((.....))))))))))...)))))............((((((......)).)))).....)))))).......... (-27.80 = -29.16 + 1.36)

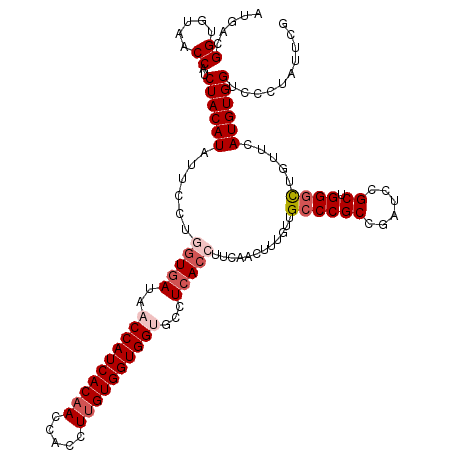
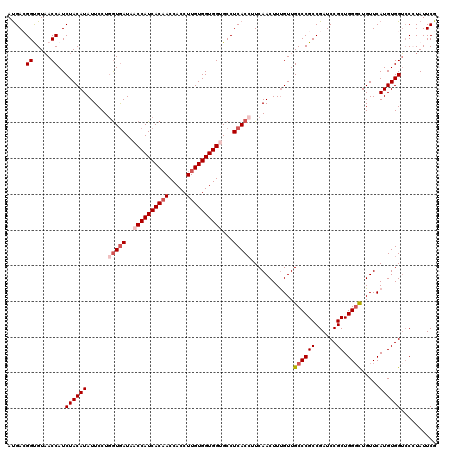
| Location | 4,222,336 – 4,222,456 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.50 |
| Mean single sequence MFE | -31.24 |
| Consensus MFE | -24.42 |
| Energy contribution | -26.22 |
| Covariance contribution | 1.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649352 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4222336 120 - 20766785 AAGAAACUCAACUCCCGCGGUCAAGUAGUCUUGUAUGGAGAUGACGGUGUAACCAUCUACAUAUUCCUGGUGAUAACCAUCACAACCACCUUGUGGUGGUGCCUCACCUUCAACUUUGUU ..(((..(((.((((......((((....))))...)))).)))..(((((......)))))......(((((..((((((((((.....))))))))))...))))))))......... ( -33.40) >DroSec_CAF1 34052 120 - 1 AAGAAAAUCAACUCCCACGGUCAAGUAGUCUUGUAUGGAGAUGACGGUGUAACCAUCUACAUAUUCCUGGUGAUAACCAUCACAACCACCUUGUGGUGGAGCCUCACCUUCAACUUUGUU ..(((..(((.((((......((((....))))...)))).)))..(((((......)))))......(((((...(((((((((.....)))))))))....))))))))......... ( -30.60) >DroSim_CAF1 61773 120 - 1 AAGAAAAUCAACUCCCACGGUCAAGUAGUCUUGUAUGGAGAUGACGGUGUAACCAUCUACAUAUUCCUGGUGAUAACCAUCACAACCACCUUGUGGUGGAGCCUCACCUUCAACUUUGUU ..(((..(((.((((......((((....))))...)))).)))..(((((......)))))......(((((...(((((((((.....)))))))))....))))))))......... ( -30.60) >DroEre_CAF1 35359 120 - 1 AAGAAGCUCAACGCCCGCGGUCAAGUAACCUUGUAUGGGGAUGAAGGCGUAACCAUCUACAUAUUCCUCGUGAUAACCAUCACCACCACCUUGUGGUGGUGCCUCACCUUCAACUUUGUU ..((((.......(((.((..((((....))))..))))).((((((.(....................(((((....)))))(((((((....)))))))...).)))))).))))... ( -32.50) >DroYak_CAF1 36589 120 - 1 AAGAAACUCAACUCCCGUGGUCAAGUAGUCUUGUACGGAGACGAAGGCGUUACCAUCUACAUAUUCCUGAUAAUAACCAUCACAACCACCUUGUGGUGGUGCCUCACAUUCAACUUCGUU ..............(((((..((((....))))))))).(((((((..((........)).......(((.....((((((((((.....))))))))))...))).......))))))) ( -29.10) >consensus AAGAAACUCAACUCCCGCGGUCAAGUAGUCUUGUAUGGAGAUGACGGUGUAACCAUCUACAUAUUCCUGGUGAUAACCAUCACAACCACCUUGUGGUGGUGCCUCACCUUCAACUUUGUU .(((...(((.((((......((((....))))...)))).))).((.....)).)))..........(((((..((((((((((.....))))))))))...)))))............ (-24.42 = -26.22 + 1.80)

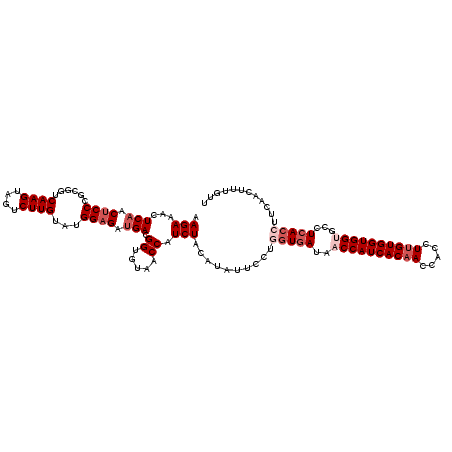
| Location | 4,222,376 – 4,222,496 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.83 |
| Mean single sequence MFE | -31.79 |
| Consensus MFE | -26.74 |
| Energy contribution | -26.74 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.988029 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4222376 120 + 20766785 AUGGUUAUCACCAGGAAUAUGUAGAUGGUUACACCGUCAUCUCCAUACAAGACUACUUGACCGCGGGAGUUGAGUUUCUUCUGUAAGACCAAUUCGAUGCCCACAUUGACUGCCAAACCU .((((..(((...((..((((.((((((........)))))).))))((((....)))).))(.(((.((((((((((((....))))..)))))))).))).)..)))..))))..... ( -32.30) >DroSec_CAF1 34092 120 + 1 AUGGUUAUCACCAGGAAUAUGUAGAUGGUUACACCGUCAUCUCCAUACAAGACUACUUGACCGUGGGAGUUGAUUUUCUUCUGCAACACCAAUUCAAUGCCCACGUUGACCACCAAACCU .((((((.(....((..((((.((((((........)))))).))))((((....)))).))(((((.(((((....................))))).)))))).))))))........ ( -31.15) >DroSim_CAF1 61813 120 + 1 AUGGUUAUCACCAGGAAUAUGUAGAUGGUUACACCGUCAUCUCCAUACAAGACUACUUGACCGUGGGAGUUGAUUUUCUUCUGCAACACCAAUUCAAUGCCCACGUUGACCACCAAACCU .((((((.(....((..((((.((((((........)))))).))))((((....)))).))(((((.(((((....................))))).)))))).))))))........ ( -31.15) >DroEre_CAF1 35399 120 + 1 AUGGUUAUCACGAGGAAUAUGUAGAUGGUUACGCCUUCAUCCCCAUACAAGGUUACUUGACCGCGGGCGUUGAGCUUCUUUUCCAAUACAAAUUCAAUGCCCACGUUGACCACCAAACCU .((((((..(((.((..((((..(((((........)))))..))))((((....)))).))..((((((((((..................)))))))))).)))))))))........ ( -34.87) >DroYak_CAF1 36629 120 + 1 AUGGUUAUUAUCAGGAAUAUGUAGAUGGUAACGCCUUCGUCUCCGUACAAGACUACUUGACCACGGGAGUUGAGUUUCUUUUCCAAUACAAAUUCAAUGCCCACGUUGACCACCAAGCCU .((((((......((..((((.((((((........)))))).))))((((....)))).))..(((.(((((((((.(........).))))))))).)))....))))))........ ( -29.50) >consensus AUGGUUAUCACCAGGAAUAUGUAGAUGGUUACACCGUCAUCUCCAUACAAGACUACUUGACCGCGGGAGUUGAGUUUCUUCUGCAACACCAAUUCAAUGCCCACGUUGACCACCAAACCU .((((..(((...((..((((.((((((........)))))).))))((((....)))).))..(((.((((((((..............)))))))).)))....)))..))))..... (-26.74 = -26.74 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:54:22 2006