| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,220,586 – 4,220,706 |
| Length | 120 |
| Max. P | 0.784305 |

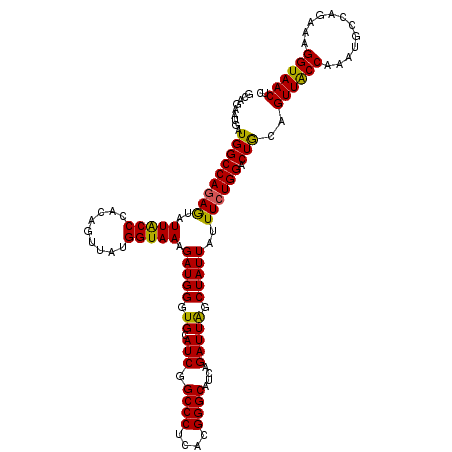
| Location | 4,220,586 – 4,220,706 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.17 |
| Mean single sequence MFE | -35.16 |
| Consensus MFE | -30.58 |
| Energy contribution | -29.82 |
| Covariance contribution | -0.76 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.762819 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4220586 120 + 20766785 GCAGAAUGAUGGCCAGAGUAUUACCCACAGUUAUGGUAAAGAUGGGUGCAUCGGCCCUCACGGGCAUCAGAUUAGCUAUUAUUUCUGGACUGCAGUUACCAAAAGCCAGAAAGGUAACUC ((((........((((((..(((((.........))))).(((((.((.(((.((((....))))....))))).)))))..)))))).))))(((((((............))))))). ( -34.30) >DroSec_CAF1 32126 120 + 1 GCAGAAUGAUGGCCAGAGUAUUACCCACAGUUAUGGUAAAGAUGGGUGCAUCGGCCCUCACGGGCAUCAGAUUAGCUAUUAUUUCUGGACUGCAGUUACCAAAAGCCAGAAAGGUAACUC ((((........((((((..(((((.........))))).(((((.((.(((.((((....))))....))))).)))))..)))))).))))(((((((............))))))). ( -34.30) >DroSim_CAF1 59438 120 + 1 GCAGAAUGAUGGCCAGAGUAUUACCCACAGUUAUGGUAAAGAUGGGUGCAUCGGCCCUCACGGGCAUCAGAUUAGCUAUUAUUUCUGGACUGCAGUUACCAAAUGCCAGAAAGGUAACUC ((((........((((((..(((((.........))))).(((((.((.(((.((((....))))....))))).)))))..)))))).))))(((((((............))))))). ( -34.30) >DroEre_CAF1 33637 120 + 1 GCAGAAUAAUGGCCAAAAUAUUACCGACAGCUAUGGUAAAGAUGGGUGCAUCGGCCCUCACGGGCAUCAGAUUGGCUAUUAUUUCUGGACUACAGUUGCCAAAUGCCAGAAAGGUAACGC .(((((((((((((((....((((((.......)))))).((((....)))).((((....))))......)))))))))).))))).......((((((............)))))).. ( -39.00) >DroYak_CAF1 34838 120 + 1 GAAGCAUUAUGGCCAGAGUAUUGCCCACAGCUAUGGUAAAGAUGGGUGCAUCGGCCCUCACGGGCAUCAGAUUGGCUAUUAUUUCUGGACUACAGUUACCAAAUGCCAGGAAGGUAACAC ((((....((((((((.....(((((.((....)).....((.((((......))))))..))))).....))))))))..)))).........((((((............)))))).. ( -33.90) >consensus GCAGAAUGAUGGCCAGAGUAUUACCCACAGUUAUGGUAAAGAUGGGUGCAUCGGCCCUCACGGGCAUCAGAUUAGCUAUUAUUUCUGGACUGCAGUUACCAAAUGCCAGAAAGGUAACUC .........(((((((((..(((((.........))))).(((((.((.(((.((((....))))....))))).)))))..)))))).)))..((((((............)))))).. (-30.58 = -29.82 + -0.76)


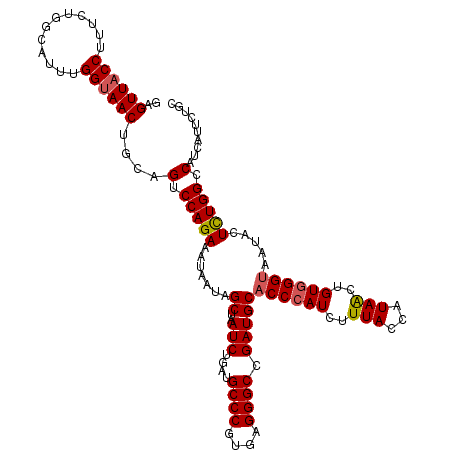
| Location | 4,220,586 – 4,220,706 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.17 |
| Mean single sequence MFE | -34.32 |
| Consensus MFE | -28.94 |
| Energy contribution | -29.14 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.784305 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4220586 120 - 20766785 GAGUUACCUUUCUGGCUUUUGGUAACUGCAGUCCAGAAAUAAUAGCUAAUCUGAUGCCCGUGAGGGCCGAUGCACCCAUCUUUACCAUAACUGUGGGUAAUACUCUGGCCAUCAUUCUGC .(((((((............)))))))((((.(((((.......((..(((....((((....)))).)))))((((((..(((...)))..)))))).....)))))........)))) ( -34.90) >DroSec_CAF1 32126 120 - 1 GAGUUACCUUUCUGGCUUUUGGUAACUGCAGUCCAGAAAUAAUAGCUAAUCUGAUGCCCGUGAGGGCCGAUGCACCCAUCUUUACCAUAACUGUGGGUAAUACUCUGGCCAUCAUUCUGC .(((((((............)))))))((((.(((((.......((..(((....((((....)))).)))))((((((..(((...)))..)))))).....)))))........)))) ( -34.90) >DroSim_CAF1 59438 120 - 1 GAGUUACCUUUCUGGCAUUUGGUAACUGCAGUCCAGAAAUAAUAGCUAAUCUGAUGCCCGUGAGGGCCGAUGCACCCAUCUUUACCAUAACUGUGGGUAAUACUCUGGCCAUCAUUCUGC .(((((((............)))))))((((.(((((.......((..(((....((((....)))).)))))((((((..(((...)))..)))))).....)))))........)))) ( -34.90) >DroEre_CAF1 33637 120 - 1 GCGUUACCUUUCUGGCAUUUGGCAACUGUAGUCCAGAAAUAAUAGCCAAUCUGAUGCCCGUGAGGGCCGAUGCACCCAUCUUUACCAUAGCUGUCGGUAAUAUUUUGGCCAUUAUUCUGC ((........(((((.(((.(....)...))))))))((((((.(((((.(((((((((....)))).((((....))))............))))).......))))).))))))..)) ( -33.70) >DroYak_CAF1 34838 120 - 1 GUGUUACCUUCCUGGCAUUUGGUAACUGUAGUCCAGAAAUAAUAGCCAAUCUGAUGCCCGUGAGGGCCGAUGCACCCAUCUUUACCAUAGCUGUGGGCAAUACUCUGGCCAUAAUGCUUC .............((((((((((....((((((((....).(((((.........((((....)))).((((....)))).........)))))))))..)))....)))).)))))).. ( -33.20) >consensus GAGUUACCUUUCUGGCAUUUGGUAACUGCAGUCCAGAAAUAAUAGCUAAUCUGAUGCCCGUGAGGGCCGAUGCACCCAUCUUUACCAUAACUGUGGGUAAUACUCUGGCCAUCAUUCUGC ..((((((............))))))....(.(((((.......((..(((....((((....)))).)))))((((((..(((...)))..)))))).....))))).).......... (-28.94 = -29.14 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:54:19 2006