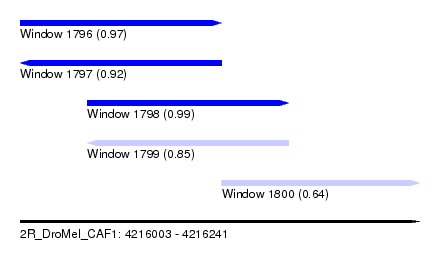
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,216,003 – 4,216,241 |
| Length | 238 |
| Max. P | 0.985196 |

| Location | 4,216,003 – 4,216,123 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.36 |
| Mean single sequence MFE | -31.60 |
| Consensus MFE | -26.77 |
| Energy contribution | -27.02 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972374 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4216003 120 + 20766785 UUAAAUGCAUUCAUUUUUCUAAACCUGGGUGUGUUUUCCACUCUUCUAUGGUCCACGACGCUGGGAUUUUUCGCCAGGCGCUCCGUGGGCAUAUUUUCUAUCGUGUUUUACUGCAUUUAC .((((((((.................(((((.(....)))))).......(((((((.((((((((....)).))).)))...))))))).....................)))))))). ( -32.00) >DroSec_CAF1 27560 120 + 1 UUAAAUGCAUUCAUUUUUUUAAACCUGGGUCUGUUUUCCACUCUUCUAUGGUCCACGACGCUUGGAUUUUUCGCCAGGCGCUCCGUGGGCAUAUUUUCCAUCGUGUUUUACUGCAUUUAC .((((((((................((((.......))))..........(((((((.(((((((........)))))))...))))))).....................)))))))). ( -32.40) >DroEre_CAF1 29337 120 + 1 UUAAAUGCAUUCAUUUUUCUGAAUCUGGGCCUGUUUUCCACACUCCUCUGGUCCACGACGCUGGGAUUUUUCGCCAGACGCUCCGUGGGCAUAUUUUGUAUCGUGUUUUACUGCAUUUAC .((((((((((((......))))...(((..(((.....)))..)))...(((((((.((((((((....)).)))).))...))))))).....................)))))))). ( -30.20) >DroYak_CAF1 30206 120 + 1 UUAAAUGCAUUCGUUUUUCUAAAUAUCGGACUGUUUUCCACACUCCUCUGGUCCACGACGCUGGGAUUUUUCGCCAGACGCUCCGUGGGCAUAUUUUCUAUCGUCUUUUACUGCAUUUAC .((((((((..................(((.(((.....))).)))....(((((((.((((((((....)).)))).))...))))))).....................)))))))). ( -31.80) >consensus UUAAAUGCAUUCAUUUUUCUAAACCUGGGUCUGUUUUCCACACUCCUAUGGUCCACGACGCUGGGAUUUUUCGCCAGACGCUCCGUGGGCAUAUUUUCUAUCGUGUUUUACUGCAUUUAC .((((((((..................((........))...........(((((((.((((((((....)).)))).))...))))))).....................)))))))). (-26.77 = -27.02 + 0.25)

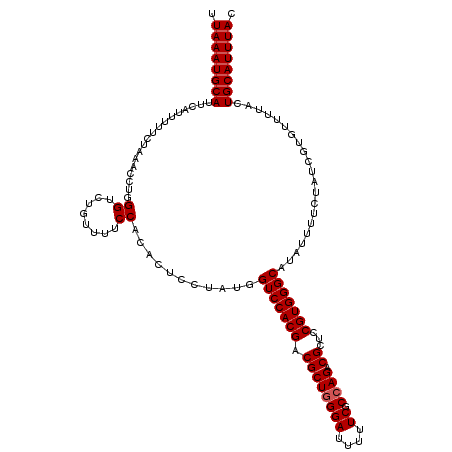
| Location | 4,216,003 – 4,216,123 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.36 |
| Mean single sequence MFE | -31.60 |
| Consensus MFE | -25.45 |
| Energy contribution | -26.20 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.923608 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4216003 120 - 20766785 GUAAAUGCAGUAAAACACGAUAGAAAAUAUGCCCACGGAGCGCCUGGCGAAAAAUCCCAGCGUCGUGGACCAUAGAAGAGUGGAAAACACACCCAGGUUUAGAAAAAUGAAUGCAUUUAA .((((((((...........((((...((((.(((((..((((.(((.((....))))))))))))))..)))).....(((.....))).......))))..........)))))))). ( -29.60) >DroSec_CAF1 27560 120 - 1 GUAAAUGCAGUAAAACACGAUGGAAAAUAUGCCCACGGAGCGCCUGGCGAAAAAUCCAAGCGUCGUGGACCAUAGAAGAGUGGAAAACAGACCCAGGUUUAAAAAAAUGAAUGCAUUUAA .((((((((((....(((..(......((((.(((((..((((.(((........))).)))))))))..))))..)..)))....))((((....))))...........)))))))). ( -29.90) >DroEre_CAF1 29337 120 - 1 GUAAAUGCAGUAAAACACGAUACAAAAUAUGCCCACGGAGCGUCUGGCGAAAAAUCCCAGCGUCGUGGACCAGAGGAGUGUGGAAAACAGGCCCAGAUUCAGAAAAAUGAAUGCAUUUAA .(((((((........................(((((..(((.((((.((....))))))))))))))......((.((.((.....)).))))..(((((......)))))))))))). ( -32.90) >DroYak_CAF1 30206 120 - 1 GUAAAUGCAGUAAAAGACGAUAGAAAAUAUGCCCACGGAGCGUCUGGCGAAAAAUCCCAGCGUCGUGGACCAGAGGAGUGUGGAAAACAGUCCGAUAUUUAGAAAAACGAAUGCAUUUAA .((((((((((.....))......((((((..(((((..(((.((((.((....))))))))))))))......(((.(((.....))).))).))))))...........)))))))). ( -34.00) >consensus GUAAAUGCAGUAAAACACGAUAGAAAAUAUGCCCACGGAGCGCCUGGCGAAAAAUCCCAGCGUCGUGGACCAGAGAAGAGUGGAAAACAGACCCAGAUUUAGAAAAAUGAAUGCAUUUAA .((((((((.......................(((((..(((.((((.((....)))))))))))))).((((......))))............................)))))))). (-25.45 = -26.20 + 0.75)

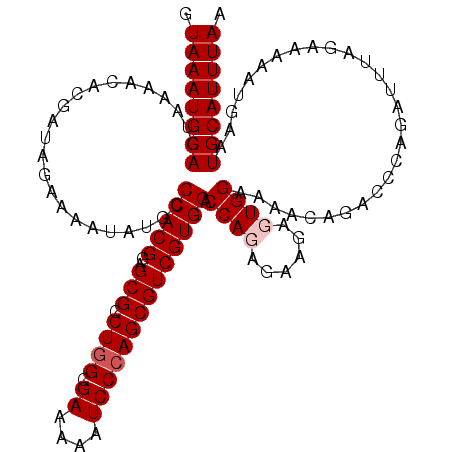
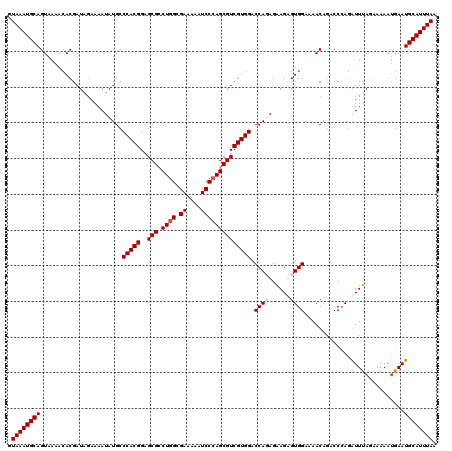
| Location | 4,216,043 – 4,216,163 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.47 |
| Mean single sequence MFE | -29.45 |
| Consensus MFE | -27.40 |
| Energy contribution | -27.65 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.985196 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4216043 120 + 20766785 CUCUUCUAUGGUCCACGACGCUGGGAUUUUUCGCCAGGCGCUCCGUGGGCAUAUUUUCUAUCGUGUUUUACUGCAUUUACUUGCUUUUUGCUAUCCUAAUUCAUAGAAAAGUCAUACAUU ((.((((((((((((((.((((((((....)).))).)))...)))))))..............((......(((......))).....))..........))))))).))......... ( -30.10) >DroSec_CAF1 27600 120 + 1 CUCUUCUAUGGUCCACGACGCUUGGAUUUUUCGCCAGGCGCUCCGUGGGCAUAUUUUCCAUCGUGUUUUACUGCAUUUACCUGCUUUUUGCUAUCCUAAUUCAUAGAAAAAUCAUACAUU ...((((((((((((((.(((((((........)))))))...)))))))............((((......))))......((.....))..........)))))))............ ( -31.60) >DroEre_CAF1 29377 120 + 1 CACUCCUCUGGUCCACGACGCUGGGAUUUUUCGCCAGACGCUCCGUGGGCAUAUUUUGUAUCGUGUUUUACUGCAUUUACCUGCUUUUUGCUAUCUUAAUUCAUAGGAAAAUCAUACACU ...((((.(((((((((.((((((((....)).)))).))...))))))).......(((..((((......)))).)))..((.....))..........)).))))............ ( -28.40) >DroYak_CAF1 30246 120 + 1 CACUCCUCUGGUCCACGACGCUGGGAUUUUUCGCCAGACGCUCCGUGGGCAUAUUUUCUAUCGUCUUUUACUGCAUUUACUUGCUUUUUGCUAUCUUAAUUCAUAGGAAAGUCAUACAUU ...((((.(((((((((.((((((((....)).)))).))...)))))))......................(((......))).................)).))))............ ( -27.70) >consensus CACUCCUAUGGUCCACGACGCUGGGAUUUUUCGCCAGACGCUCCGUGGGCAUAUUUUCUAUCGUGUUUUACUGCAUUUACCUGCUUUUUGCUAUCCUAAUUCAUAGAAAAAUCAUACAUU ...((((((((((((((.((((((((....)).)))).))...)))))))......................(((......))).................)))))))............ (-27.40 = -27.65 + 0.25)

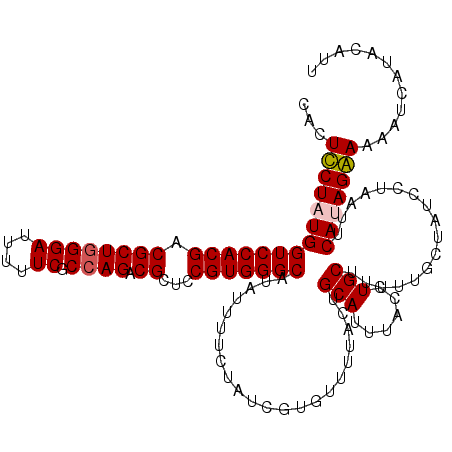
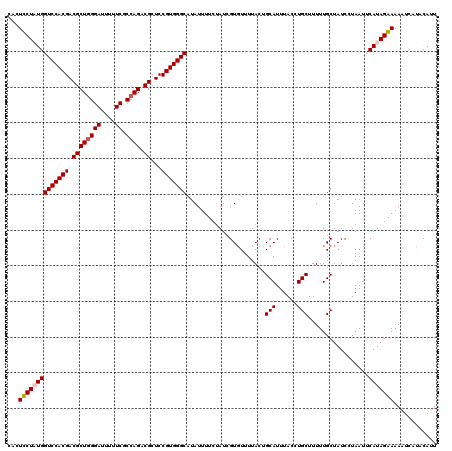
| Location | 4,216,043 – 4,216,163 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.47 |
| Mean single sequence MFE | -28.85 |
| Consensus MFE | -27.70 |
| Energy contribution | -28.20 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.852522 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4216043 120 - 20766785 AAUGUAUGACUUUUCUAUGAAUUAGGAUAGCAAAAAGCAAGUAAAUGCAGUAAAACACGAUAGAAAAUAUGCCCACGGAGCGCCUGGCGAAAAAUCCCAGCGUCGUGGACCAUAGAAGAG .........((((((((((..........(((....(((......))).((.....))...........)))(((((..((((.(((.((....))))))))))))))..)))))))))) ( -31.50) >DroSec_CAF1 27600 120 - 1 AAUGUAUGAUUUUUCUAUGAAUUAGGAUAGCAAAAAGCAGGUAAAUGCAGUAAAACACGAUGGAAAAUAUGCCCACGGAGCGCCUGGCGAAAAAUCCAAGCGUCGUGGACCAUAGAAGAG ..........(((((((((..........(((....(((......))).((.....))...........)))(((((..((((.(((........))).)))))))))..))))))))). ( -29.10) >DroEre_CAF1 29377 120 - 1 AGUGUAUGAUUUUCCUAUGAAUUAAGAUAGCAAAAAGCAGGUAAAUGCAGUAAAACACGAUACAAAAUAUGCCCACGGAGCGUCUGGCGAAAAAUCCCAGCGUCGUGGACCAGAGGAGUG ...........(((((.((..........(((....(((......))).(((........)))......)))(((((..(((.((((.((....))))))))))))))..)).))))).. ( -28.30) >DroYak_CAF1 30246 120 - 1 AAUGUAUGACUUUCCUAUGAAUUAAGAUAGCAAAAAGCAAGUAAAUGCAGUAAAAGACGAUAGAAAAUAUGCCCACGGAGCGUCUGGCGAAAAAUCCCAGCGUCGUGGACCAGAGGAGUG ...........(((((.((..........(((....(((......))).((.....))...........)))(((((..(((.((((.((....))))))))))))))..)).))))).. ( -26.50) >consensus AAUGUAUGACUUUCCUAUGAAUUAAGAUAGCAAAAAGCAAGUAAAUGCAGUAAAACACGAUAGAAAAUAUGCCCACGGAGCGCCUGGCGAAAAAUCCCAGCGUCGUGGACCAGAGAAGAG .........((((((((((..........(((....(((......))).((.....))...........)))(((((..(((.((((.((....))))))))))))))..)))))))))) (-27.70 = -28.20 + 0.50)

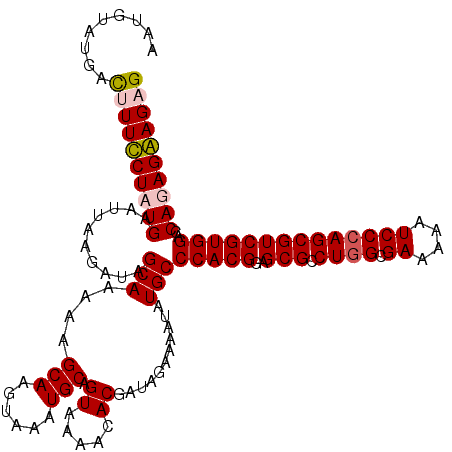
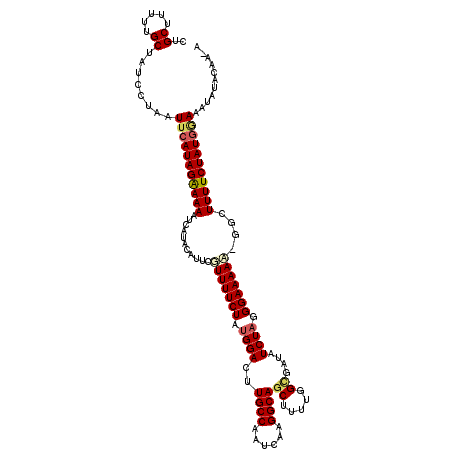
| Location | 4,216,123 – 4,216,241 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.27 |
| Mean single sequence MFE | -26.90 |
| Consensus MFE | -19.99 |
| Energy contribution | -20.86 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.643574 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4216123 118 + 20766785 UUGCUUUUUGCUAUCCUAAUUCAUAGAAAAGUCAUACAUUCGUUUUCUGUGGACUUGCCAGUCAAGGCAGCAUUCGGUGAUAUCUAGGGAAAAA-G-GUUUUCUAUGGAAAUAUACAAAA (..((...((((.......(((((((((((............)))))))))))...(((......)))))))...))..)..((((.(((((..-.-..))))).))))........... ( -25.10) >DroSec_CAF1 27680 118 + 1 CUGCUUUUUGCUAUCCUAAUUCAUAGAAAAAUCAUACAUUCGUUUUCUUCGGACCUGCCAGUCACGGCAGCUUUUGGCGAUAUCUAGGGAAAAA-GGGUUUUCUAUGGAAAUAUACAA-A ..((.....))........((((((((((........((((.((((((..(((..((((......))))((.....))....)))..)))))).-)))))))))))))).........-. ( -26.00) >DroEre_CAF1 29457 118 + 1 CUGCUUUUUGCUAUCUUAAUUCAUAGGAAAAUCAUACACUCCUUUUCUAUGGACUUGCCCAUCAAGGCAGCCUUUGGAGACAUCUAGGGAAAAG-GGCUUUUCUAUUUAUAUAUGCAA-A .(((.((((.((((........)))).))))........(((((((((.((((.(((((......)))))....((....)))))).)))))))-)).................))).-. ( -26.50) >DroYak_CAF1 30326 119 + 1 UUGCUUUUUGCUAUCUUAAUUCAUAGGAAAGUCAUACAUUCGUUUUCUAUGGACAUGCCAAUCAAGGCAGCUUUUGGCGAUAUCUAGGGAAAACAUACUUUUCUAUGAAAAUAUACAA-A ..((.....))........(((((((((((((.((......(((((((.((((..((((......))))((.....))....)))).)))))))))))))))))))))).........-. ( -30.00) >consensus CUGCUUUUUGCUAUCCUAAUUCAUAGAAAAAUCAUACAUUCGUUUUCUAUGGACUUGCCAAUCAAGGCAGCUUUUGGCGAUAUCUAGGGAAAAA_GGCUUUUCUAUGGAAAUAUACAA_A ..((.....))........(((((((((((...........(((((((.((((..((((......))))((.....))....)))).)))))))....)))))))))))........... (-19.99 = -20.86 + 0.87)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:54:13 2006