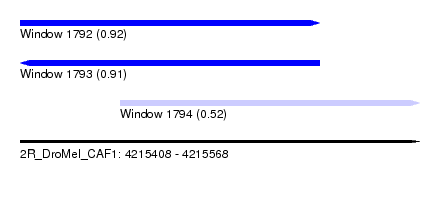
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,215,408 – 4,215,568 |
| Length | 160 |
| Max. P | 0.924803 |

| Location | 4,215,408 – 4,215,528 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.61 |
| Mean single sequence MFE | -39.65 |
| Consensus MFE | -31.94 |
| Energy contribution | -34.00 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924803 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4215408 120 + 20766785 UCCUCCCAUAGCUUUGCUAUCAUCUAGGGGCAACUCGCUGUGGUACGUUGCCAUGGGCGAGGAAGCCAUUUAUGCUAUGUAUUCCCUGCCCAUAACCGUGCCGAUAGCGGUCUUUAUGUU ......(((((..((((((((.....(((....))).....((((((.....(((((((.((((..(((.......)))..)))).)))))))...))))))))))))))...))))).. ( -44.00) >DroSec_CAF1 26970 120 + 1 UCUUCCCAUAGCUUUGCUCAGCUCUAGGGGCAACUCGCUGUGGUACGUUGCCAUGGCCGAGGAAUCCAUUUAUGCUGUGUAUUCCCUGCCUAUAACCAUGCCGAUAGCAGUCUUUAUGUU ...((((..((((......))))...))))..(((.(((((((((.((((....(((.(.(((((.(((.......))).)))))).)))..))))..)))).))))))))......... ( -32.00) >DroEre_CAF1 28745 119 + 1 -CCUCCGAUAGCUUUGUUGAGCUCUAGGGGCGAUUCGCUGUGGUACGUUGCCAUGGGCGAGGAAUUCAUGUAUGCAGUGUAUUCCCUGCCCAUAACCGUGCCAAUAGCGGUUUUUAUGUU -.((((...(((((....)))))...))))....(((((((((((((.....(((((((.(((((.(((.......))).))))).)))))))...)))))).))))))).......... ( -46.40) >DroYak_CAF1 29607 120 + 1 UCCUCCGAUAGCUUUGUUAAGCUCUAGGGGCACAUCGCUGUGGUACGUUGCCAUGACCGAGGAAUACAUUUAUGCAGUGUAUUCCCUGCCCAUAACCAUGCCAAUAGCGGUUUUUAUGUU ..((((...(((((....)))))...))))...((((((((((((.(((...(((..((.((((((((((.....)))))))))).))..))))))..)))).))))))))......... ( -36.20) >consensus UCCUCCCAUAGCUUUGCUAAGCUCUAGGGGCAACUCGCUGUGGUACGUUGCCAUGGCCGAGGAAUCCAUUUAUGCAGUGUAUUCCCUGCCCAUAACCAUGCCAAUAGCGGUCUUUAUGUU ..((((...(((((....)))))...))))....(((((((((((((.....(((((((.(((((.(((.......))).))))).)))))))...)))))).))))))).......... (-31.94 = -34.00 + 2.06)

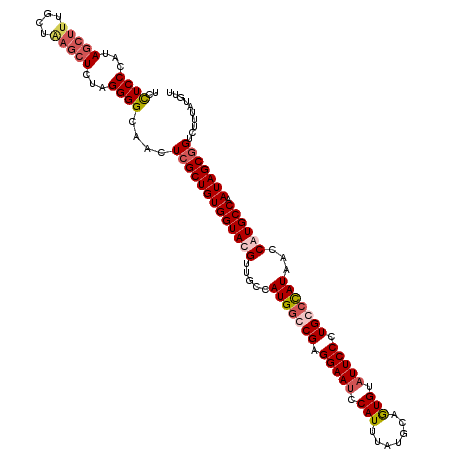
| Location | 4,215,408 – 4,215,528 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.61 |
| Mean single sequence MFE | -38.30 |
| Consensus MFE | -28.45 |
| Energy contribution | -30.33 |
| Covariance contribution | 1.87 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.905283 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4215408 120 - 20766785 AACAUAAAGACCGCUAUCGGCACGGUUAUGGGCAGGGAAUACAUAGCAUAAAUGGCUUCCUCGCCCAUGGCAACGUACCACAGCGAGUUGCCCCUAGAUGAUAGCAAAGCUAUGGGAGGA ........(((((((...((.((((((((((((..((((..(((.......)))..))))..)))))))))..))).))..)))).)))...(((...(.(((((...))))).).))). ( -41.80) >DroSec_CAF1 26970 120 - 1 AACAUAAAGACUGCUAUCGGCAUGGUUAUAGGCAGGGAAUACACAGCAUAAAUGGAUUCCUCGGCCAUGGCAACGUACCACAGCGAGUUGCCCCUAGAGCUGAGCAAAGCUAUGGGAAGA ........(((((((...((.((((((((.(((.((((((...((.......)).))))))..))))))))..))).))..))).))))..(((...((((......))))..))).... ( -33.10) >DroEre_CAF1 28745 119 - 1 AACAUAAAAACCGCUAUUGGCACGGUUAUGGGCAGGGAAUACACUGCAUACAUGAAUUCCUCGCCCAUGGCAACGUACCACAGCGAAUCGCCCCUAGAGCUCAACAAAGCUAUCGGAGG- ...........((((..(((.((((((((((((..(((((.((.((....)))).)))))..)))))))))..))).))).)))).....((((...((((......))))...)).))- ( -40.70) >DroYak_CAF1 29607 120 - 1 AACAUAAAAACCGCUAUUGGCAUGGUUAUGGGCAGGGAAUACACUGCAUAAAUGUAUUCCUCGGUCAUGGCAACGUACCACAGCGAUGUGCCCCUAGAGCUUAACAAAGCUAUCGGAGGA ..........((((((..(((((.(((((((.(..((((((((.(.....).))))))))..).)))))))..(((......)))..)))))..)))(((((....)))))..))).... ( -37.60) >consensus AACAUAAAAACCGCUAUCGGCACGGUUAUGGGCAGGGAAUACACAGCAUAAAUGGAUUCCUCGCCCAUGGCAACGUACCACAGCGAGUUGCCCCUAGAGCUUAACAAAGCUAUCGGAGGA ...........((((...((.((((((((((((..(((((.((.........)).)))))..)))))))))..))).))..)))).....((((((.((((......)))).)))).)). (-28.45 = -30.33 + 1.87)

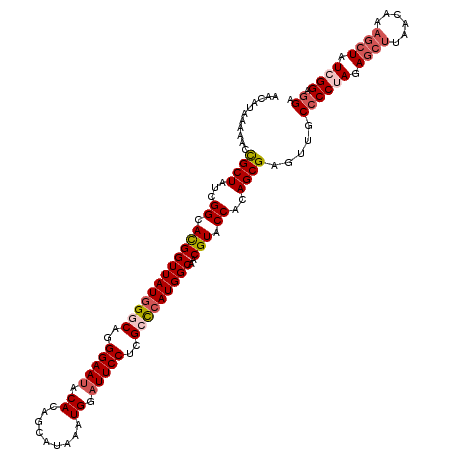
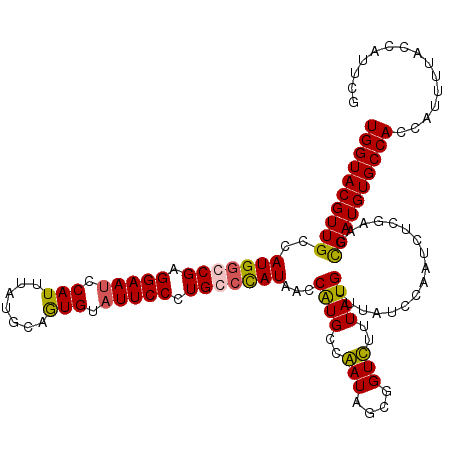
| Location | 4,215,448 – 4,215,568 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.08 |
| Mean single sequence MFE | -31.93 |
| Consensus MFE | -27.06 |
| Energy contribution | -26.75 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.85 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.520210 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4215448 120 + 20766785 UGGUACGUUGCCAUGGGCGAGGAAGCCAUUUAUGCUAUGUAUUCCCUGCCCAUAACCGUGCCGAUAGCGGUCUUUAUGUUUAUCCAAUCUCGAACCGAUGUGCCACCAUUUUACCAUUCG ((((((((((..(((((((.((((..(((.......)))..)))).))))))).(((((.......)))))........................))))))))))............... ( -35.90) >DroSec_CAF1 27010 120 + 1 UGGUACGUUGCCAUGGCCGAGGAAUCCAUUUAUGCUGUGUAUUCCCUGCCUAUAACCAUGCCGAUAGCAGUCUUUAUGUUUAUCCAAUCUCGAACCGAUGUGCCACCAUUUUACCAUUCG ((((((((((....(((.(.(((((.(((.......))).)))))).))).((((.((((..(((....)))..)))).))))............))))))))))............... ( -27.80) >DroEre_CAF1 28784 120 + 1 UGGUACGUUGCCAUGGGCGAGGAAUUCAUGUAUGCAGUGUAUUCCCUGCCCAUAACCGUGCCAAUAGCGGUUUUUAUGUUUUUCCAAUCGCGAACUGAUGUGCCACCAUUUUACCACUCG (((((((((...(((((((.(((((.(((.......))).))))).)))))))((((((.......))))))........................)))))))))............... ( -36.00) >DroYak_CAF1 29647 120 + 1 UGGUACGUUGCCAUGACCGAGGAAUACAUUUAUGCAGUGUAUUCCCUGCCCAUAACCAUGCCAAUAGCGGUUUUUAUGUUUAUCCAAUCCCGAACCGAUGUGCCACCAUUUUACCAUUCG ((((((((((..........((((((((((.....)))))))))).....(((((....(((......)))..))))).................))))))))))............... ( -28.00) >consensus UGGUACGUUGCCAUGGCCGAGGAAUCCAUUUAUGCAGUGUAUUCCCUGCCCAUAACCAUGCCAAUAGCGGUCUUUAUGUUUAUCCAAUCUCGAACCGAUGUGCCACCAUUUUACCAUUCG ((((((((((..(((((((.(((((.(((.......))).))))).)))))))...((((..(((....)))..)))).................))))))))))............... (-27.06 = -26.75 + -0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:54:08 2006