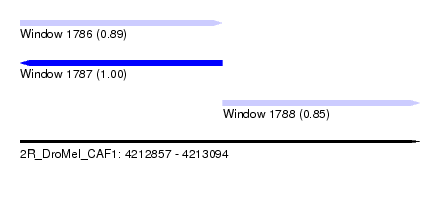
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,212,857 – 4,213,094 |
| Length | 237 |
| Max. P | 0.998239 |

| Location | 4,212,857 – 4,212,977 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.58 |
| Mean single sequence MFE | -44.88 |
| Consensus MFE | -33.70 |
| Energy contribution | -34.74 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4212857 120 + 20766785 CGAGUAAUCAAUCCUCGCCCCGCGUCCAGAUGGUCCACAACCUGGGCGGCAUAGACCUCCAGGCUGGCCGUAUAGAUCACCAGGUCGUACCACUUGGACACAAAGUCCAGGAACUCGUCU (((((......((((.((...))((((((.((((..((.(((((((((((.(((.((....))))))))))........)))))).)))))).)))))).........)))))))))... ( -42.70) >DroSec_CAF1 24515 120 + 1 CGAGUAAUCAAUCCUCGCCCCGCGUCCAAUUGGUCCACAACCUGGGCGGCAUAAGCCUCCAGGCUGGCCGUGUAGAUCACCAGGUCGUACCACUUGGACACGAAGUCCAGGAACUCGUCA (((((......((((.((..((.((((((.((((..((.(((((((((((...((((....)))).)))))(.....).)))))).)))))).)))))).))..).).)))))))))... ( -50.20) >DroSim_CAF1 56458 120 + 1 CGAGUAAUCAAUCCUCGCCCCGCGUCCAGUUGGUCCACAACCUGGGCGGCAUAAGCCUCCAGGCUGGCCGUGUAGAUCACCAGGUCGUACCACUUGGACACGAAGUCCAGGAACUCGUCG (((((......((((.((..((.((((((.((((..((.(((((((((((...((((....)))).)))))(.....).)))))).)))))).)))))).))..).).)))))))))... ( -49.90) >DroEre_CAF1 26376 120 + 1 CGAGACAUCCUUCCCUGUCCCGCGUCCAGGAGGUCCACCACCUGGGCAGCAUAGACCUCCAGACUGGCCGUGUAGACCACCAGGUCGUACCACUUGGACACGAAGUGCAGGAACUCAUCG .(((...((((.(.((....((.(((((((.(((..((.((((((.(.(((..(.((........)))..))).)....)))))).))))).))))))).)).)).).)))).))).... ( -41.60) >DroYak_CAF1 27195 120 + 1 CGAGACAUUCUUCCUUGUCCCGCGUCCAGAAGGUCCACCACCUGGGUGGCAUAGACCUCCAGGCUGGCCGUGUAGAUCACCAGGUCGUACCACUUGGACACGCAGUCCAGGAACUCAUCG .(((((..............((((.((((..((....)).((((((.((......)))))))))))).))))..((((....))))))....(((((((.....)))))))..))).... ( -40.00) >consensus CGAGUAAUCAAUCCUCGCCCCGCGUCCAGAUGGUCCACAACCUGGGCGGCAUAGACCUCCAGGCUGGCCGUGUAGAUCACCAGGUCGUACCACUUGGACACGAAGUCCAGGAACUCGUCG .((((......((((.....((.((((((.((((..((.(((((((((((...(.((....)).).)))))........)))))).)))))).)))))).))......)))))))).... (-33.70 = -34.74 + 1.04)

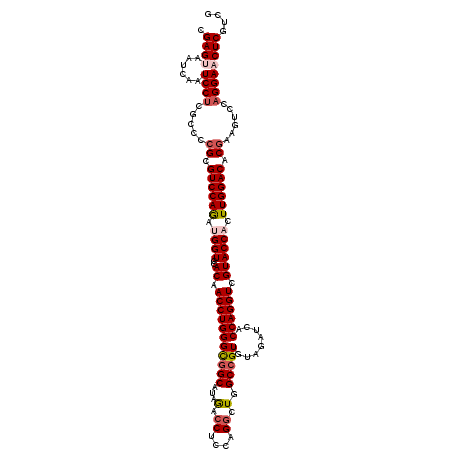
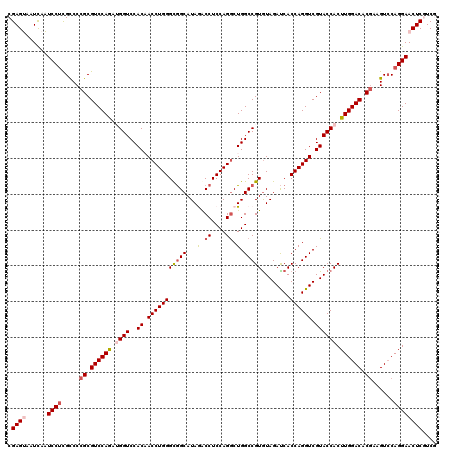
| Location | 4,212,857 – 4,212,977 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.58 |
| Mean single sequence MFE | -54.66 |
| Consensus MFE | -45.86 |
| Energy contribution | -46.66 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.84 |
| SVM decision value | 3.04 |
| SVM RNA-class probability | 0.998239 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4212857 120 - 20766785 AGACGAGUUCCUGGACUUUGUGUCCAAGUGGUACGACCUGGUGAUCUAUACGGCCAGCCUGGAGGUCUAUGCCGCCCAGGUUGUGGACCAUCUGGACGCGGGGCGAGGAUUGAUUACUCG ...(((((((((.(.(((((((((((..(((((((((((((.(...((((.((((........))))))))...))))))))))..))))..)))))))))))).))))......))))) ( -55.30) >DroSec_CAF1 24515 120 - 1 UGACGAGUUCCUGGACUUCGUGUCCAAGUGGUACGACCUGGUGAUCUACACGGCCAGCCUGGAGGCUUAUGCCGCCCAGGUUGUGGACCAAUUGGACGCGGGGCGAGGAUUGAUUACUCG ...(((((((((.(.((((((((((((.(((((((((((((((.....))((((.((((....))))...)))).)))))))))..)))).))))))))))))).))))......))))) ( -59.50) >DroSim_CAF1 56458 120 - 1 CGACGAGUUCCUGGACUUCGUGUCCAAGUGGUACGACCUGGUGAUCUACACGGCCAGCCUGGAGGCUUAUGCCGCCCAGGUUGUGGACCAACUGGACGCGGGGCGAGGAUUGAUUACUCG ...(((((((((.(.(((((((((((..(((((((((((((((.....))((((.((((....))))...)))).)))))))))..))))..)))))))))))).))))......))))) ( -58.90) >DroEre_CAF1 26376 120 - 1 CGAUGAGUUCCUGCACUUCGUGUCCAAGUGGUACGACCUGGUGGUCUACACGGCCAGUCUGGAGGUCUAUGCUGCCCAGGUGGUGGACCUCCUGGACGCGGGACAGGGAAGGAUGUCUCG ....(((.((((...(((((((((((........(((.((((.((....)).))))))).((((((((..((..(....)..)))))))))))))))))))...))...))))...))). ( -52.40) >DroYak_CAF1 27195 120 - 1 CGAUGAGUUCCUGGACUGCGUGUCCAAGUGGUACGACCUGGUGAUCUACACGGCCAGCCUGGAGGUCUAUGCCACCCAGGUGGUGGACCUUCUGGACGCGGGACAAGGAAGAAUGUCUCG .((((..((((((..((((((((((....)).))...(((((..........)))))((.((((((((..(((((....))))))))))))).))))))))..).)))))...))))... ( -47.20) >consensus CGACGAGUUCCUGGACUUCGUGUCCAAGUGGUACGACCUGGUGAUCUACACGGCCAGCCUGGAGGUCUAUGCCGCCCAGGUUGUGGACCAACUGGACGCGGGGCGAGGAUUGAUUACUCG ...(((((((((.(.(((((((((((.(((((.(.(((((((..........)))))(((((.(((....)))..)))))..)).)))))).)))))))))))).))))......))))) (-45.86 = -46.66 + 0.80)

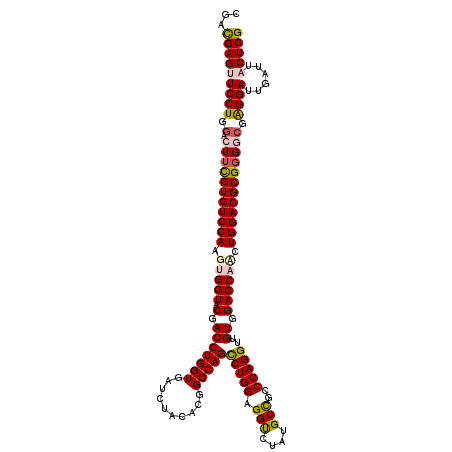
| Location | 4,212,977 – 4,213,094 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.57 |
| Mean single sequence MFE | -46.89 |
| Consensus MFE | -34.72 |
| Energy contribution | -35.20 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.849176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4212977 117 + 20766785 ACAUGCGGCCGCUUGAAGACCCGGAAAACGAUAGGUUC---CAUCCCGUCGAUGGAAAUAUUGAGCACAUAGUCCGGUUGUGCGUGUUCGGGCAACUGGCUACAUCCCACGUUGUCGUGG ..(((..((((.(((....((((((...(((((..(((---((((.....))))))).))))).(((((.........)))))...))))))))).))))..))).(((((....))))) ( -43.10) >DroSec_CAF1 24635 117 + 1 ACAUGCGGCCGCUUGAAGACCCGGAAAACGAUAGGUUC---CGUCCCGUCGAUGGAAAUAUUGAGCACAUAGUCCGGUCGUGCGUUUUCGGGCAACUGGCUGCAUCCCACGUUAUCGUGG ..(((((((((.(((....((((((((.(((((..(((---((((.....))))))).))))).((((...........)))).))))))))))).))))))))).(((((....))))) ( -47.90) >DroSim_CAF1 56578 117 + 1 ACAUGCGGCCGCUUGAAGACCCGGAAAACGAUAGGUUC---CAUCCCGUCGAUGGAAAUAUUGAGCACAUAGUCCGGUCGUGCGUUUUCGGGCAACUGGCUGCAUCCCACGUUAUCGUGG ..(((((((((.(((....((((((((.(((((..(((---((((.....))))))).))))).((((...........)))).))))))))))).))))))))).(((((....))))) ( -48.30) >DroEre_CAF1 26496 120 + 1 ACAUGCGGCCGCUUGAAGACCCGAAACACGAUCGGUUCGGACAUGGGUUCGAUGGAAACGUUGAGCACAUAAUCCGGUUGCGCGUGAUCGGGCAGCUGGCUGCAACCCACGUUGUCAUGG ...((((((((((....(.(((((..((((......(((((.(((.((((((((....)))))))).)))..))))).....)))).))))))))).)))))))..(((.(....).))) ( -48.70) >DroYak_CAF1 27315 120 + 1 ACAUGCGGCCGCUUGAAGACCCGAAACACGAUCGGCUCGACCAUCGGAUCGAUGGACACGUUGAGUACAUAGUCCGGCUGGGCGUGAUCGGGCAACUGGCUGCAACCCACGUUGUCAUGG ...((((((((.(((....(((((..((((..(((((.(((.......((((((....)))))).......))).)))))..)))).)))))))).))))))))..(((.(....).))) ( -46.44) >consensus ACAUGCGGCCGCUUGAAGACCCGGAAAACGAUAGGUUC___CAUCCCGUCGAUGGAAAUAUUGAGCACAUAGUCCGGUUGUGCGUGUUCGGGCAACUGGCUGCAUCCCACGUUGUCGUGG ...((((((((.(((....((((.(((((...................((((((....))))))((((...........)))))))))))))))).))))))))..(((((....))))) (-34.72 = -35.20 + 0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:54:02 2006