| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,212,067 – 4,212,183 |
| Length | 116 |
| Max. P | 0.808025 |

| Location | 4,212,067 – 4,212,183 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.66 |
| Mean single sequence MFE | -37.82 |
| Consensus MFE | -28.70 |
| Energy contribution | -29.70 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.768468 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4212067 116 + 20766785 UUCUCCGAACUUCUGCUCACACACGGAAAUCGCAAAAAAGCCAG----CCGGGCCAACCUGGCGUAUGAGCAAUGAAAUGCAUCUGCUCUGAGGUUGGCACAUAUGUGCCUGCUUUCGCC .....((((.(((((........)))))...(((....((((.(----(((((....))))))....((((((((.....))).)))))...))))(((((....)))))))).)))).. ( -38.20) >DroSec_CAF1 23760 116 + 1 UUCCCGGAACUUCUGCUCACACACGGAAAUCGCAAAAAAGGCAA----CCGGGCCAACCUGGCGUAUGAGCAAUGAAAUGCAUCUGCUCCGAGGUUGGCCCAUAUGUGCCUGCCUUCGCC ....((((..(((((........)))))..........(((((.----..((((((((((((.(((.(((((......))).))))).)).)))))))))).....)))))...)))).. ( -41.30) >DroSim_CAF1 55665 116 + 1 UUCCCCAAACUUUUGCUCACACACGGAAAUCGCAAAAAAGCCAG----CCGGGCCAACCUGGCGUAUGAGCAAUGAAAUGCAUCUGCUCCGAGGUUGGCCCAUAUGUGCCUGCCUUCGCC ..............((...((((.((.....((......))...----))((((((((((((.(((.(((((......))).))))).)).))))))))))...))))...))....... ( -38.00) >DroEre_CAF1 25672 115 + 1 UUCCCCGAGCUUCUGCUCACACACGGAAAUCGCAAAAAAGCCAG----CCGGCCC-ACCUGGCGUAUGAGCAAUAAAACGCAUCUGCUGCGAGGUUGGCCCAUAUGUGCCUGCUUUCGUC ......((((....))))....((((((...(((.....((((.----..((((.-((((.(((((.((((........)).))))).)).)))).))))....)).)).))))))))). ( -35.60) >DroYak_CAF1 26385 119 + 1 UUCCCCGAACUUCUGCUCACACACGGAAAUCGCAAAAAAGCCAGCCAGCCGGGCC-ACCUGGCGUAUGAGCAAUAAAAUGCAUCUGCUCUGGGGUUGGCCCAUAUGUGCCUGCUUUCAUC .....(((..(((((........))))).)))....(((((..((((...(((((-(((..(.(((.(((((......))).))))).)..))..))))))...)).))..))))).... ( -36.00) >consensus UUCCCCGAACUUCUGCUCACACACGGAAAUCGCAAAAAAGCCAG____CCGGGCCAACCUGGCGUAUGAGCAAUGAAAUGCAUCUGCUCCGAGGUUGGCCCAUAUGUGCCUGCUUUCGCC ..............((...((((.((.....((......)).......))((((((((((((.(((.(((((......))).))))).)).))))))))))...))))...))....... (-28.70 = -29.70 + 1.00)

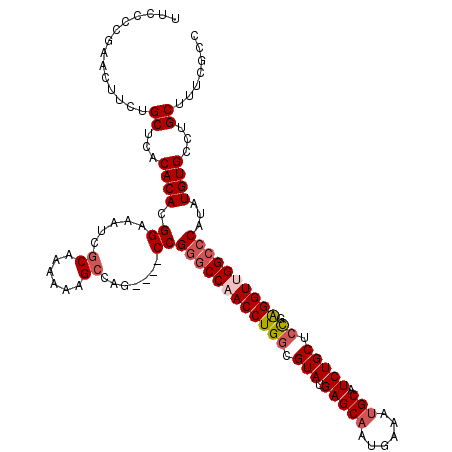
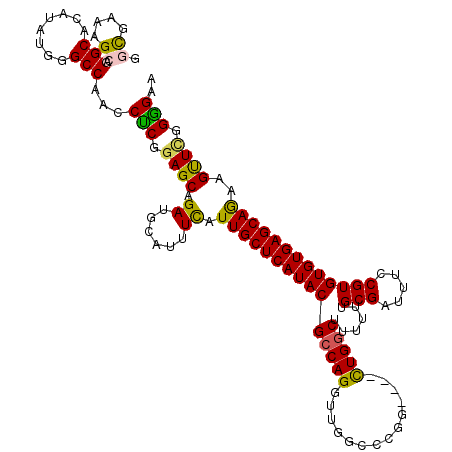
| Location | 4,212,067 – 4,212,183 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.66 |
| Mean single sequence MFE | -44.49 |
| Consensus MFE | -34.50 |
| Energy contribution | -33.98 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808025 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4212067 116 - 20766785 GGCGAAAGCAGGCACAUAUGUGCCAACCUCAGAGCAGAUGCAUUUCAUUGCUCAUACGCCAGGUUGGCCCGG----CUGGCUUUUUUGCGAUUUCCGUGUGUGAGCAGAAGUUCGGAGAA .((....)).(((((....)))))...(((.((((.((......)).(((((((((((((.((....)).))----)..((......)).........))))))))))..)))).))).. ( -46.50) >DroSec_CAF1 23760 116 - 1 GGCGAAGGCAGGCACAUAUGGGCCAACCUCGGAGCAGAUGCAUUUCAUUGCUCAUACGCCAGGUUGGCCCGG----UUGCCUUUUUUGCGAUUUCCGUGUGUGAGCAGAAGUUCCGGGAA .(((((((.(((((....(((((((((((.((.......(((......))).......))))))))))))).----.))))))))))))..((((((.....((((....)))))))))) ( -48.54) >DroSim_CAF1 55665 116 - 1 GGCGAAGGCAGGCACAUAUGGGCCAACCUCGGAGCAGAUGCAUUUCAUUGCUCAUACGCCAGGUUGGCCCGG----CUGGCUUUUUUGCGAUUUCCGUGUGUGAGCAAAAGUUUGGGGAA .((....)).(((........)))..(((((((((....))......(((((((((((((.((....)).))----)..((......)).........))))))))))...))))))).. ( -41.80) >DroEre_CAF1 25672 115 - 1 GACGAAAGCAGGCACAUAUGGGCCAACCUCGCAGCAGAUGCGUUUUAUUGCUCAUACGCCAGGU-GGGCCGG----CUGGCUUUUUUGCGAUUUCCGUGUGUGAGCAGAAGCUCGGGGAA (.(....)).(((........)))..(((((((.....)))(((((..((((((((((((((.(-(...)).----)))))......(((.....)))))))))))))))))..)))).. ( -41.30) >DroYak_CAF1 26385 119 - 1 GAUGAAAGCAGGCACAUAUGGGCCAACCCCAGAGCAGAUGCAUUUUAUUGCUCAUACGCCAGGU-GGCCCGGCUGGCUGGCUUUUUUGCGAUUUCCGUGUGUGAGCAGAAGUUCGGGGAA ..........(((........)))..((((.(((((((.....))).(((((((((((((((.(-(((...)))).)))))......(((.....)))))))))))))..)))))))).. ( -44.30) >consensus GGCGAAAGCAGGCACAUAUGGGCCAACCUCGGAGCAGAUGCAUUUCAUUGCUCAUACGCCAGGUUGGCCCGG____CUGGCUUUUUUGCGAUUUCCGUGUGUGAGCAGAAGUUCGGGGAA .((....)).(((........)))...(((.((((.((......)).(((((((((((((((..............)))))......(((.....)))))))))))))..)))).))).. (-34.50 = -33.98 + -0.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:53:59 2006