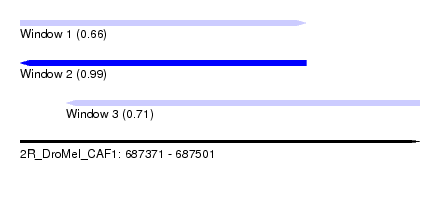
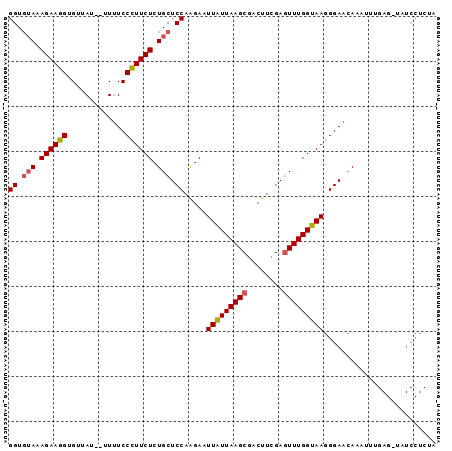
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 687,371 – 687,501 |
| Length | 130 |
| Max. P | 0.988244 |

| Location | 687,371 – 687,464 |
|---|---|
| Length | 93 |
| pre Sequences | 3 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 74.20 |
| Mean single sequence MFE | -15.87 |
| Consensus MFE | -12.25 |
| Energy contribution | -11.59 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.659709 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 687371 93 + 20766785 UAGAGGAUAACUCAAAUUUUUUCCCUUGCCAAACUCAAAGUAGCUUAACAAUUCUUGGAGCAGAGAAAGGAAAUAUAUAACACUUUCUUU--ACC ..(((.....))).............................((((...........))))((((((((.............))))))))--... ( -12.32) >DroSec_CAF1 15700 93 + 1 UAGAGGAUAGCUCAAAUUUGUUCCCUUACCAAACUCGAAGUCGCUUAAUAAUUCCUGGAGCAGAGAAGGGAUAA--AUAACCCCUUCUUUACACC ..(((.....)))......(((((...........((....)).............)))))(((((((((....--.....)))))))))..... ( -18.75) >DroSim_CAF1 2893 76 + 1 -----------------UUGUUCCCUUACCAAAAUCGAAGUCGCUUAAUAACUCUUGGAGCAGAGAAGGGAAAA--AUAGCACCUUCUCUACACC -----------------..(((((...........((....)).............)))))((((((((.....--......))))))))..... ( -16.55) >consensus UAGAGGAUA_CUCAAAUUUGUUCCCUUACCAAACUCGAAGUCGCUUAAUAAUUCUUGGAGCAGAGAAGGGAAAA__AUAACACCUUCUUUACACC ..........................................((((...........))))((((((((.............))))))))..... (-12.25 = -11.59 + -0.66)

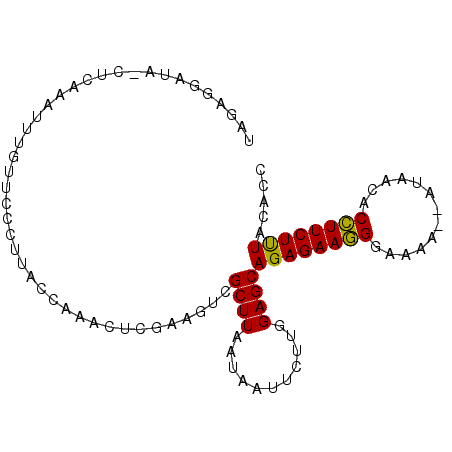
| Location | 687,371 – 687,464 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 74.20 |
| Mean single sequence MFE | -23.40 |
| Consensus MFE | -15.37 |
| Energy contribution | -15.49 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.66 |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.988244 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 687371 93 - 20766785 GGU--AAAGAAAGUGUUAUAUAUUUCCUUUCUCUGCUCCAAGAAUUGUUAAGCUACUUUGAGUUUGGCAAGGGAAAAAAUUUGAGUUAUCCUCUA .((--(.((((((.(..........))))))).)))(((.....((((((((((......)))))))))).)))........(((.....))).. ( -18.90) >DroSec_CAF1 15700 93 - 1 GGUGUAAAGAAGGGGUUAU--UUAUCCCUUCUCUGCUCCAGGAAUUAUUAAGCGACUUCGAGUUUGGUAAGGGAACAAAUUUGAGCUAUCCUCUA ((.(((.(((((((((...--..))))))))).))).))((((.(((((((((........)))))))))(....)............))))... ( -29.50) >DroSim_CAF1 2893 76 - 1 GGUGUAGAGAAGGUGCUAU--UUUUCCCUUCUCUGCUCCAAGAGUUAUUAAGCGACUUCGAUUUUGGUAAGGGAACAA----------------- ((.((((((((((......--.....)))))))))).)).....(((((((.((....))...)))))))(....)..----------------- ( -21.80) >consensus GGUGUAAAGAAGGUGUUAU__UUUUCCCUUCUCUGCUCCAAGAAUUAUUAAGCGACUUCGAGUUUGGUAAGGGAACAAAUUUGAG_UAUCCUCUA ((.(((.((((((.............)))))).))).)).....(((((((((........)))))))))......................... (-15.37 = -15.49 + 0.11)

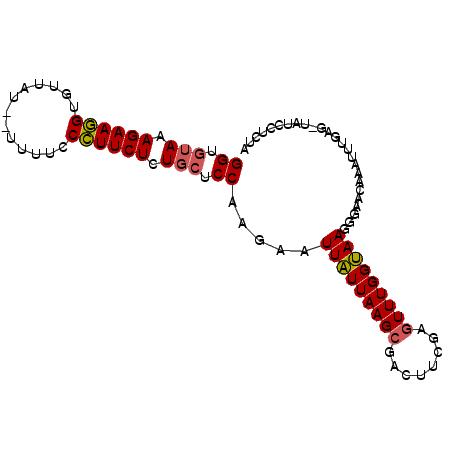
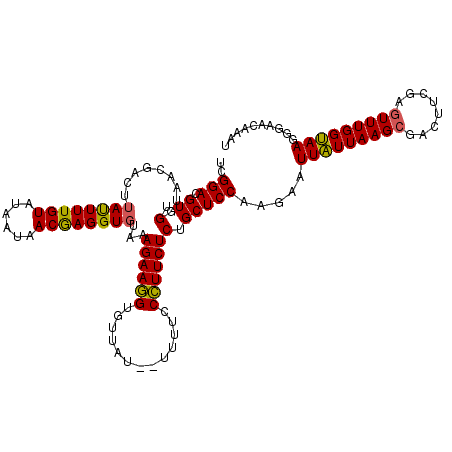
| Location | 687,386 – 687,501 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 82.23 |
| Mean single sequence MFE | -31.40 |
| Consensus MFE | -19.21 |
| Energy contribution | -18.55 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713077 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 687386 115 - 20766785 UCGGACGUUGGUAACGAUUUACUUUGUAUAAUAACAAGGU--AAAGAAAGUGUUAUAUAUUUCCUUUCUCUGCUCCAAGAAUUGUUAAGCUACUUUGAGUUUGGCAAGGGAAAAAAU ..(((.....((((((.((((((((((......)))))))--))).....)))))).....))).........(((.....((((((((((......)))))))))).)))...... ( -28.60) >DroSec_CAF1 15715 109 - 1 UCGGACGUUGG------CUUAUUUUGUGUAAUAACGAGGUGUAAAGAAGGGGUUAU--UUAUCCCUUCUCUGCUCCAGGAAUUAUUAAGCGACUUCGAGUUUGGUAAGGGAACAAAU (((((.((((.------((((..((((......))))((.(((.(((((((((...--..))))))))).))).)).........)))))))))))))(((((.........))))) ( -32.70) >DroSim_CAF1 2893 113 - 1 UCGGACGUUGGUAACGACUUAUUUUGUAUAAUAACGAGGUGUAGAGAAGGUGCUAU--UUUUCCCUUCUCUGCUCCAAGAGUUAUUAAGCGACUUCGAUUUUGGUAAGGGAACAA-- (((((.((((....)))).....((((.(((((((..((.((((((((((......--.....)))))))))).))....))))))).)))).))))).........(....)..-- ( -32.90) >consensus UCGGACGUUGGUAACGACUUAUUUUGUAUAAUAACGAGGUGUAAAGAAGGUGUUAU__UUUUCCCUUCUCUGCUCCAAGAAUUAUUAAGCGACUUCGAGUUUGGUAAGGGAACAAAU ..(((.((.(.........((((((((......))))))))...((((((.............))))))).))))).....(((((((((........))))))))).......... (-19.21 = -18.55 + -0.66)

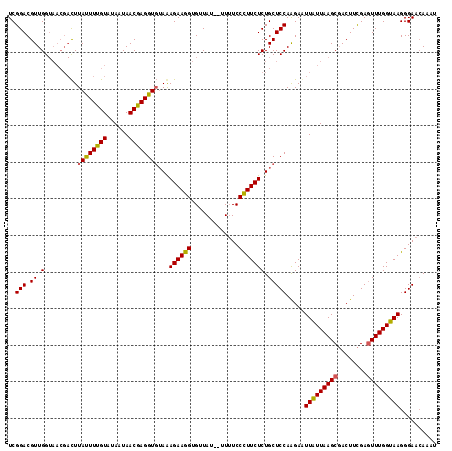
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:23:02 2006