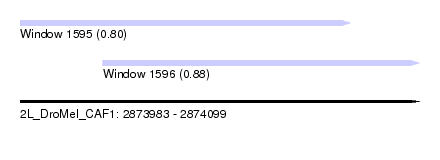
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,873,983 – 2,874,099 |
| Length | 116 |
| Max. P | 0.876903 |

| Location | 2,873,983 – 2,874,079 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 85.61 |
| Mean single sequence MFE | -21.96 |
| Consensus MFE | -18.98 |
| Energy contribution | -18.90 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.796794 |
| Prediction | RNA |
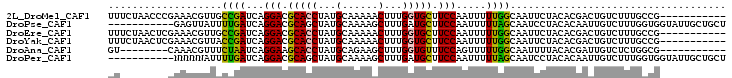
Download alignment: ClustalW | MAF
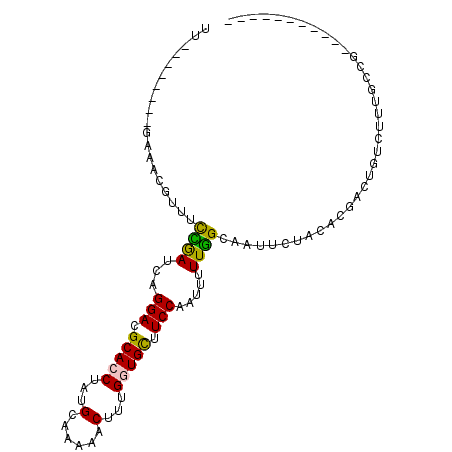
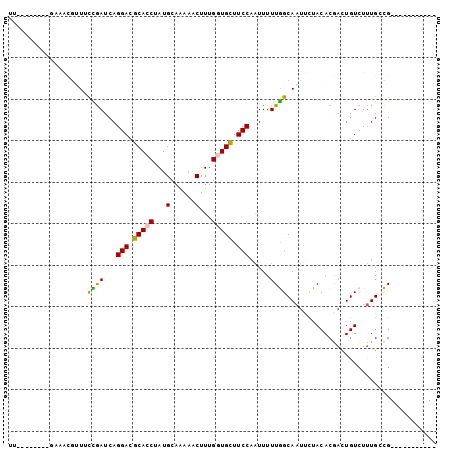
>2L_DroMel_CAF1 2873983 96 + 22407834 GCUGUCAAUUUUUUAAUC----UUUGUCUUUCUAACCCGAAACGUUGCCGAUCAGGACGCACCUAUGCAAAAACUUUGGUGCUUCCAAUUUUUGGCAAUU ..................----......((((......)))).((((((((...(((.(((((..............))))).))).....)))))))). ( -21.44) >DroSec_CAF1 8856 96 + 1 GCCGUCAGUUUUUUAAUC----UUUGUCUUUCUAACUCGAAACGUUGCCGAUCAGGACGCACCUAUGCAAAAACUUUGGUGCUUCCAAUUUUUGGCAAUU ..................----......((((......)))).((((((((...(((.(((((..............))))).))).....)))))))). ( -21.44) >DroSim_CAF1 9237 95 + 1 GCCGUCAGUUU-UUAAUC----UUUGUCUUUCUAACUCGAAACGUUGCCGAUCAGGACGCACCUAUGCAAAAACUUUGGUGCUUCCAAUUUUUGGCAAUU ...........-......----......((((......)))).((((((((...(((.(((((..............))))).))).....)))))))). ( -21.44) >DroEre_CAF1 9284 96 + 1 GCUAUCUGGUUUUUAAUC----UUUGUCUUUCUAACUCGAAACGUUGCCGAUCAGGACGCACCUAUGCAAAAACUUUGGUGCUUCCAAUUUUUGGCAAUU ....((.((((.......----...........)))).))...((((((((...(((.(((((..............))))).))).....)))))))). ( -21.91) >DroYak_CAF1 9239 96 + 1 GCUGUCUGGUUUUUAAUC----UUUGUCUUUCUAACUCGAAACGUUACCGAUCAGGACGCACCUAUGCAAAAACUUUGGUGCUUCCAAUUUUUGGCAAUU ..(((((((((..((((.----((((...........))))..))))..)))))(((.(((((..............))))).))).......))))... ( -18.34) >DroAna_CAF1 9193 84 + 1 -------GUUU-UCGAUUGGGGUUUGUUGU--------CAAACGUUUCUAAUCAGGAAGCACCUAUGCAGAAGCUUUGGUGUUUCCAGUUUUUGGCAAUU -------....-..((((((((((((....--------)))))...))))))).(((((((((...((....))...))))))))).............. ( -27.20) >consensus GCUGUCAGUUUUUUAAUC____UUUGUCUUUCUAACUCGAAACGUUGCCGAUCAGGACGCACCUAUGCAAAAACUUUGGUGCUUCCAAUUUUUGGCAAUU ...........................................((((((((...(((.(((((...(......)...))))).))).....)))))))). (-18.98 = -18.90 + -0.08)

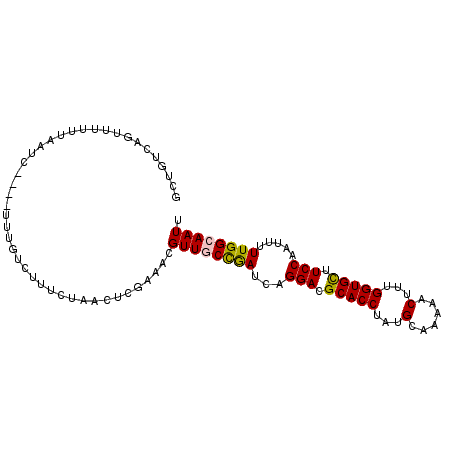
| Location | 2,874,007 – 2,874,099 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 73.90 |
| Mean single sequence MFE | -22.62 |
| Consensus MFE | -11.74 |
| Energy contribution | -11.10 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.876903 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2874007 92 + 22407834 UUUCUAACCCGAAACGUUGCCGAUCAGGACGCACCUAUGCAAAAACUUUGGUGCUUCCAAUUUUUGGCAAUUCUACACGACUGUCUUUGCCG----------- ((((......)))).((((((((...(((.(((((..............))))).))).....)))))))).....................----------- ( -21.34) >DroPse_CAF1 10207 92 + 1 -----------GAGUUAUUUUGAUCAGGACGCAGCUAUGCAAAAGCUUUGAUGCUUCCAAUUUUUAGCAAUCCUACACAAUUGUCUUUGGUGGUAUUGCUGCU -----------...............(((.(((.....((....)).....))).)))......(((((((.((((.(((......))))))).))))))).. ( -23.10) >DroEre_CAF1 9308 92 + 1 UUUCUAACUCGAAACGUUGCCGAUCAGGACGCACCUAUGCAAAAACUUUGGUGCUUCCAAUUUUUGGCAAUUCUACACGACUGUCUUUGCCG----------- ........(((....((((((((...(((.(((((..............))))).))).....))))))))......)))............----------- ( -21.94) >DroYak_CAF1 9263 92 + 1 UUUCUAACUCGAAACGUUACCGAUCAGGACGCACCUAUGCAAAAACUUUGGUGCUUCCAAUUUUUGGCAAUUCUACACGACUGUCUUUGCCG----------- ((((......))))............(((.(((((..............))))).))).......(((((....(((....)))..))))).----------- ( -19.44) >DroAna_CAF1 9213 84 + 1 GU--------CAAACGUUUCUAAUCAGGAAGCACCUAUGCAGAAGCUUUGGUGUUUCCAGUUUUUGGCAAUUUUACACGAUUGUCUCUGGCG----------- ((--------((..............(((((((((...((....))...))))))))).......(((((((......)))))))..)))).----------- ( -26.80) >DroPer_CAF1 10186 92 + 1 -----------NNNNNAUUUUGAUCAGGACGCAGCUAUGCAAAAGCUUUGAUGCUUCCAAUUUUUAGCAAUCCUACACAAUUGUCUUUGGUGGUAUUGCUGCU -----------...............(((.(((.....((....)).....))).)))......(((((((.((((.(((......))))))).))))))).. ( -23.10) >consensus UU________GAAACGUUUCCGAUCAGGACGCACCUAUGCAAAAACUUUGGUGCUUCCAAUUUUUGGCAAUUCUACACGACUGUCUUUGCCG___________ ...................((((...(((.(((((...(......)...))))).))).....)))).................................... (-11.74 = -11.10 + -0.64)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:34 2006