| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,858,059 – 2,858,153 |
| Length | 94 |
| Max. P | 0.991714 |

| Location | 2,858,059 – 2,858,153 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 71.83 |
| Mean single sequence MFE | -23.26 |
| Consensus MFE | -14.45 |
| Energy contribution | -15.28 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.62 |
| SVM decision value | 2.28 |
| SVM RNA-class probability | 0.991714 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
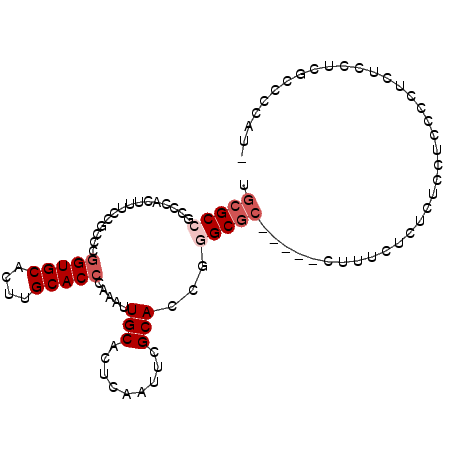
>2L_DroMel_CAF1 2858059 94 + 22407834 UGCGCCGCCCACUUUCCGCCCGGUGCACUUGCACCCAAAUUGCACUCAAUUUGCACCGGGCGC-----CUUUCCCUUUCCUCCGCUCUCCACGCCCCAU- .(((............(((((((((((..((((.......)))).......))))))))))).-----..............)))..............- ( -25.43) >DroPse_CAF1 17910 92 + 1 UGCGC---CUACGUUCGACUCGGUGCAGCUGCACCUAAAUUGCACUCAACGCGCACUCAGCGC-----CAAGCGAACGCCUCCCCUCCCGUCACUCCCUC .....---...((((((.((.(((((....)))))...............((((.....))))-----..))))))))...................... ( -23.10) >DroSec_CAF1 8545 94 + 1 UGCGCCGCCCACUUUCCGCACGGUGCACUUGCACCCAAAUUGCACUCAAUUUGCACCGGGCGC-----CUUUCUCUCUCCUCCGCUCUCCUCGCCCCAU- .(((((((.........)).(((((((..((((.......)))).......))))))))))))-----...............................- ( -23.20) >DroSim_CAF1 8472 94 + 1 UGCGCCGCCCACUUUCCGCCCGGUGCACUUGCACCCAAAUUGCACUCAAUUUGCACCGGGCGC-----CUUUCUCUCUCCUCCGCUCUCCUUGCCCCAU- .(((............(((((((((((..((((.......)))).......))))))))))).-----..............)))..............- ( -25.43) >DroAna_CAF1 7535 94 + 1 UGCGCCACCCACAACACGCCCAGUGCAAGUGCACCCAUGUUGCACCCAAU-CGCACUUGGCCAACGAAUUGUGUCUUUGCCCCCCU----UUGCCCCCU- .(((.....(((((...(((.(((((..(((((.......))))).....-.))))).))).......)))))....)))......----.........- ( -19.30) >DroPer_CAF1 16961 92 + 1 UGCGC---CUACGUUCGACUCGGUGCAGCUGCACCUAAAUUGCACUCAACGCGCACUCAGCGC-----CAAGCGAACGCCUCCCCUCCCGUCACUCCCUC .....---...((((((.((.(((((....)))))...............((((.....))))-----..))))))))...................... ( -23.10) >consensus UGCGCCGCCCACUUUCCGCCCGGUGCACUUGCACCCAAAUUGCACUCAAUUCGCACCGGGCGC_____CUUUCUCUCUCCUCCCCUCUCCUCGCCCCAU_ .(((((...............(((((....))))).....(((.........)))...)))))..................................... (-14.45 = -15.28 + 0.83)

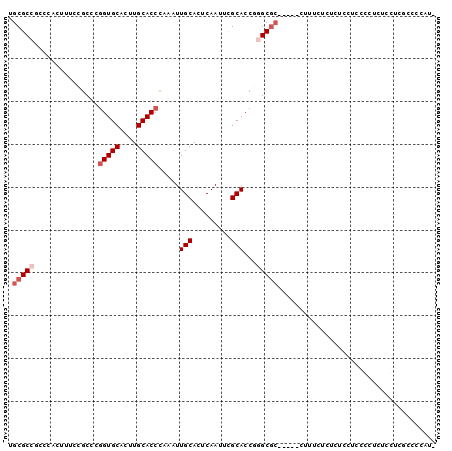
| Location | 2,858,059 – 2,858,153 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 71.83 |
| Mean single sequence MFE | -31.50 |
| Consensus MFE | -18.64 |
| Energy contribution | -18.92 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.876944 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
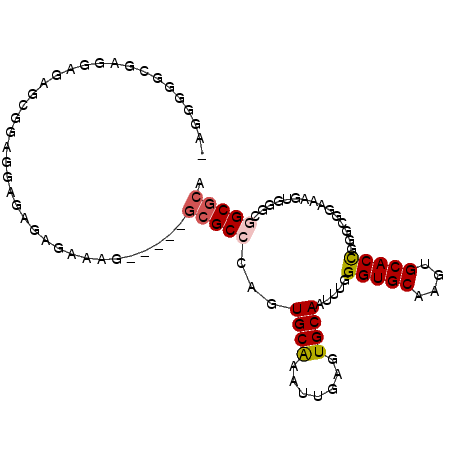
>2L_DroMel_CAF1 2858059 94 - 22407834 -AUGGGGCGUGGAGAGCGGAGGAAAGGGAAAG-----GCGCCCGGUGCAAAUUGAGUGCAAUUUGGGUGCAAGUGCACCGGGCGGAAAGUGGGCGGCGCA -.....((((.(...((...............-----.(((((((((((.......((((.......))))..)))))))))))....))...).)))). ( -33.65) >DroPse_CAF1 17910 92 - 1 GAGGGAGUGACGGGAGGGGAGGCGUUCGCUUG-----GCGCUGAGUGCGCGUUGAGUGCAAUUUAGGUGCAGCUGCACCGAGUCGAACGUAG---GCGCA ......(((.(....).....(((((((((((-----...((((((((((.....)))).))))))((((....)))))))).)))))))..---.))). ( -32.00) >DroSec_CAF1 8545 94 - 1 -AUGGGGCGAGGAGAGCGGAGGAGAGAGAAAG-----GCGCCCGGUGCAAAUUGAGUGCAAUUUGGGUGCAAGUGCACCGUGCGGAAAGUGGGCGGCGCA -((.(((((.......(........)......-----.))))).)).(((((((....)))))))(((((....)))))((((.(........).)))). ( -31.44) >DroSim_CAF1 8472 94 - 1 -AUGGGGCAAGGAGAGCGGAGGAGAGAGAAAG-----GCGCCCGGUGCAAAUUGAGUGCAAUUUGGGUGCAAGUGCACCGGGCGGAAAGUGGGCGGCGCA -..............(((..............-----.(((((((((((.......((((.......))))..)))))))))))....(....)..))). ( -30.40) >DroAna_CAF1 7535 94 - 1 -AGGGGGCAA----AGGGGGGCAAAGACACAAUUCGUUGGCCAAGUGCG-AUUGGGUGCAACAUGGGUGCACUUGCACUGGGCGUGUUGUGGGUGGCGCA -.........----....(.((.....((((((....((.(((.(((((-....((((((.......)))))))))))))).)).))))))....)).). ( -29.50) >DroPer_CAF1 16961 92 - 1 GAGGGAGUGACGGGAGGGGAGGCGUUCGCUUG-----GCGCUGAGUGCGCGUUGAGUGCAAUUUAGGUGCAGCUGCACCGAGUCGAACGUAG---GCGCA ......(((.(....).....(((((((((((-----...((((((((((.....)))).))))))((((....)))))))).)))))))..---.))). ( -32.00) >consensus _AGGGGGCGAGGAGAGCGGAGGAGAGAGAAAG_____GCGCCCAGUGCAAAUUGAGUGCAAUUUGGGUGCAAGUGCACCGGGCGGAAAGUGGGCGGCGCA .....................................(((((...((((.......)))).....(((((....)))))...............))))). (-18.64 = -18.92 + 0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:25 2006