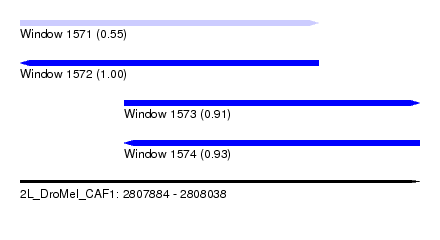
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,807,884 – 2,808,038 |
| Length | 154 |
| Max. P | 0.996483 |

| Location | 2,807,884 – 2,807,999 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.94 |
| Mean single sequence MFE | -34.91 |
| Consensus MFE | -25.22 |
| Energy contribution | -24.62 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.554366 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2807884 115 + 22407834 GAGUGCCAAAAAUGCCAACAGAUAGUGACGCCGCCAACUGG-GGGCGUGGCAUUGUCAGGGUGGCG----GGAGGUUAAAAAUAAAAUUGCAAUUAAGAAUUUUAUGCAAUUCGGUGGCU ....((((....(((((.(.((((((((((((.((....))-.)))))..))))))).)..)))))----...............(((((((.............)))))))...)))). ( -36.42) >DroSec_CAF1 30775 119 + 1 GAGUGCCAAAAAUGCCAUCAGAUAGUGACGCCGCCCACUGG-GGGCGUGGCAUCGUCAGGUUGGGGAGGGGGAGGUUAAAAAUAAAAGUGCAAUUAAGAAUUUUAUGCAAUUCGGUGGCU .............((((((.(((.((.(((((.(((...))-)))))).)))))...........................................(((((......))))))))))). ( -30.50) >DroSim_CAF1 32621 119 + 1 GAGUGCCAAAAAUGCCAUCAGAUAGUGACGCCGCCCACUGG-GGGCGUGGCAUCGUCAGGUUGGGGAGGUGGAGGUUAAAAAUAAAAGUGCAAUUAAGAAUUUUAUGCAAUUCGGUGGCU ....((((......(((((.....((.(((((.(((...))-)))))).)).((.((.....)).))))))).........................(((((......)))))..)))). ( -32.90) >DroEre_CAF1 31478 108 + 1 GGGUGCCAAAAAAGCAGCCAGAUAGUGACGCCGCCCAUUGG-GGGCGUGGCAUCGUUGGGU-----------GGGCUAAAAAUAAAACUGCAAUUCAGAAUUUUAUGCAAUUCGGUGGCU ....(((.....(((.(((.(((.((.(((((.(((...))-)))))).)))))....)))-----------..))).........((((.((((((........)).))))))))))). ( -34.80) >DroYak_CAF1 30658 109 + 1 GAGCGCCAAAAAUGCCGCCACAUAGUGACGCCGCCAAUGGGGGGGCGUGGCAUGGUGGGGU-----------GGGUUAAAAAUAAAACUGCAAUUAAGAAUUUUAUGCAAUUUGGCGGCU .(((((((((....(((((.(((.((.(((((.((.....)).))))).))...))).)))-----------))..............((((.............)))).)))))).))) ( -39.92) >consensus GAGUGCCAAAAAUGCCACCAGAUAGUGACGCCGCCCACUGG_GGGCGUGGCAUCGUCAGGUUGG_G____GGAGGUUAAAAAUAAAACUGCAAUUAAGAAUUUUAUGCAAUUCGGUGGCU .............((((((.(((.((.(((((.((.....).)))))).)))))..................................((((.............))))....)))))). (-25.22 = -24.62 + -0.60)

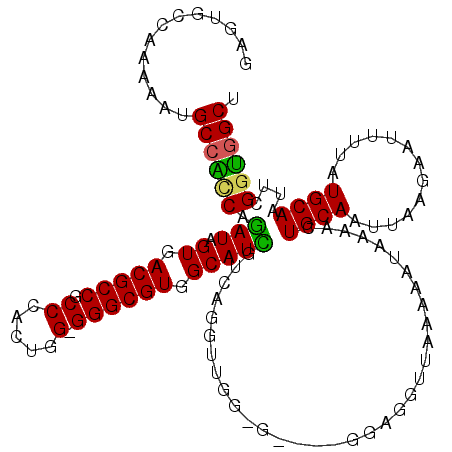
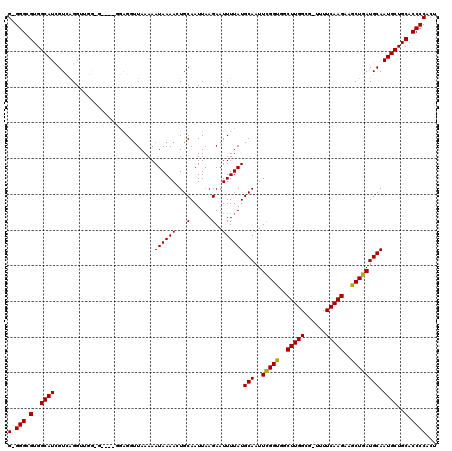
| Location | 2,807,884 – 2,807,999 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.94 |
| Mean single sequence MFE | -29.96 |
| Consensus MFE | -25.56 |
| Energy contribution | -25.84 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.70 |
| SVM RNA-class probability | 0.996483 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2807884 115 - 22407834 AGCCACCGAAUUGCAUAAAAUUCUUAAUUGCAAUUUUAUUUUUAACCUCC----CGCCACCCUGACAAUGCCACGCCC-CCAGUUGGCGGCGUCACUAUCUGUUGGCAUUUUUGGCACUC .((((..((((((((.............))))))))..............----.((((.(..((.(.((..(((((.-((....)).))))))).).)).).)))).....)))).... ( -28.82) >DroSec_CAF1 30775 119 - 1 AGCCACCGAAUUGCAUAAAAUUCUUAAUUGCACUUUUAUUUUUAACCUCCCCCUCCCCAACCUGACGAUGCCACGCCC-CCAGUGGGCGGCGUCACUAUCUGAUGGCAUUUUUGGCACUC .((((..(((((......)))))...........................................((((((((((((-((...))).))))(((.....))))))))))..)))).... ( -28.00) >DroSim_CAF1 32621 119 - 1 AGCCACCGAAUUGCAUAAAAUUCUUAAUUGCACUUUUAUUUUUAACCUCCACCUCCCCAACCUGACGAUGCCACGCCC-CCAGUGGGCGGCGUCACUAUCUGAUGGCAUUUUUGGCACUC .((((..(((((......)))))...........................................((((((((((((-((...))).))))(((.....))))))))))..)))).... ( -28.00) >DroEre_CAF1 31478 108 - 1 AGCCACCGAAUUGCAUAAAAUUCUGAAUUGCAGUUUUAUUUUUAGCCC-----------ACCCAACGAUGCCACGCCC-CCAAUGGGCGGCGUCACUAUCUGGCUGCUUUUUUGGCACCC .((((..((((((((.............))))))))......(((((.-----------.......((((((..((((-.....)))))))))).......)))))......)))).... ( -33.18) >DroYak_CAF1 30658 109 - 1 AGCCGCCAAAUUGCAUAAAAUUCUUAAUUGCAGUUUUAUUUUUAACCC-----------ACCCCACCAUGCCACGCCCCCCCAUUGGCGGCGUCACUAUGUGGCGGCAUUUUUGGCGCUC (((.(((((((((((.............))))))..............-----------........(((((..(((((......)).)))(((((...))))))))))..)))))))). ( -31.82) >consensus AGCCACCGAAUUGCAUAAAAUUCUUAAUUGCACUUUUAUUUUUAACCUCC____C_CCAACCUGACGAUGCCACGCCC_CCAGUGGGCGGCGUCACUAUCUGAUGGCAUUUUUGGCACUC .((((..((((((((.............))))))))..............................((((((.((((........))))..((((.....))))))))))..)))).... (-25.56 = -25.84 + 0.28)

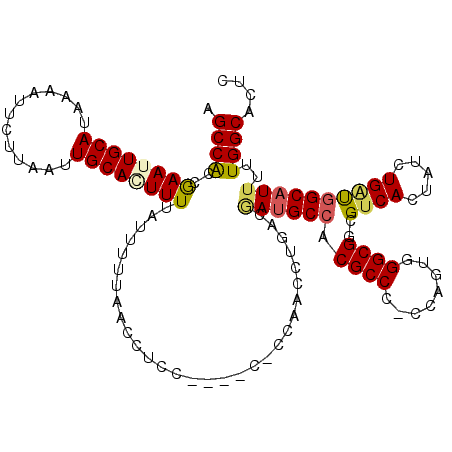
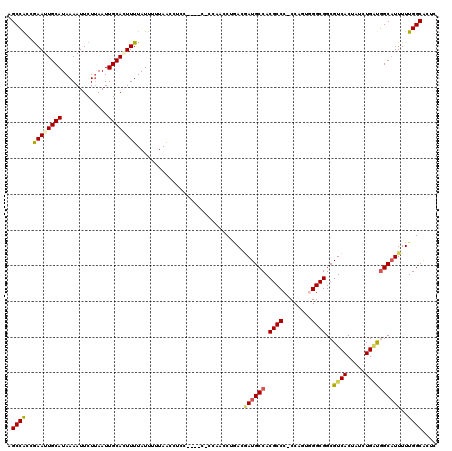
| Location | 2,807,924 – 2,808,038 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.45 |
| Mean single sequence MFE | -36.69 |
| Consensus MFE | -28.42 |
| Energy contribution | -28.10 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908465 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2807924 114 + 22407834 G-GGGCGUGGCAUUGUCAGGGUGGCG----GGAGGUUAAAAAUAAAAUUGCAAUUAAGAAUUUUAUGCAAUUCGGUGGCUUGGCG-UUUUCAAGAAGCUGAUGCAAUGCUGCACCCCACU (-(((.(..(((((((.......(((----.((((((...(((.........)))...)))))).)))...(((((..(((((..-...)))))..))))).)))))))..).))))... ( -37.20) >DroSec_CAF1 30815 118 + 1 G-GGGCGUGGCAUCGUCAGGUUGGGGAGGGGGAGGUUAAAAAUAAAAGUGCAAUUAAGAAUUUUAUGCAAUUCGGUGGCUUGGCG-UUUUCAAGAAGCUGAUGCAAUGCUGCACCCCACU (-(((.(..((((((((((.((......(..((.(((((..((.....((((.............)))).....))...))))).-))..)...)).))))))..))))..).))))... ( -36.42) >DroSim_CAF1 32661 118 + 1 G-GGGCGUGGCAUCGUCAGGUUGGGGAGGUGGAGGUUAAAAAUAAAAGUGCAAUUAAGAAUUUUAUGCAAUUCGGUGGCUUGGCG-UUUUCAAGAAGCUGAUGCAAUGCUGCACCCCACU (-(((.(..((((((((((.(((..((.((.(((.(((..........((((.............))))......)))))).)).-))..)))....))))))..))))..).))))... ( -37.31) >DroEre_CAF1 31518 107 + 1 G-GGGCGUGGCAUCGUUGGGU-----------GGGCUAAAAAUAAAACUGCAAUUCAGAAUUUUAUGCAAUUCGGUGGCUUGGCG-UUUUCAAGAAGCUGAUGCAAUGCUGCACCCCACU (-(((.(..((((..((((((-----------..((.............)).)))))).......((((..(((((..(((((..-...)))))..)))))))))))))..).))))... ( -36.62) >DroYak_CAF1 30698 109 + 1 GGGGGCGUGGCAUGGUGGGGU-----------GGGUUAAAAAUAAAACUGCAAUUAAGAAUUUUAUGCAAUUUGGCGGCUUGGCGAUUUUCAAGUAGCUGAUGCAAUGCUGCACCCCACU .((((.(..((((..((.(((-----------...(((....))).))).)).............((((..(..((.((((((......)))))).))..)))))))))..).))))... ( -35.90) >consensus G_GGGCGUGGCAUCGUCAGGUUGG_G____GGAGGUUAAAAAUAAAACUGCAAUUAAGAAUUUUAUGCAAUUCGGUGGCUUGGCG_UUUUCAAGAAGCUGAUGCAAUGCUGCACCCCACU (.(((.(..((((............................((((((...(......)..))))))(((..(((((..(((((......)))))..)))))))).))))..).))))... (-28.42 = -28.10 + -0.32)

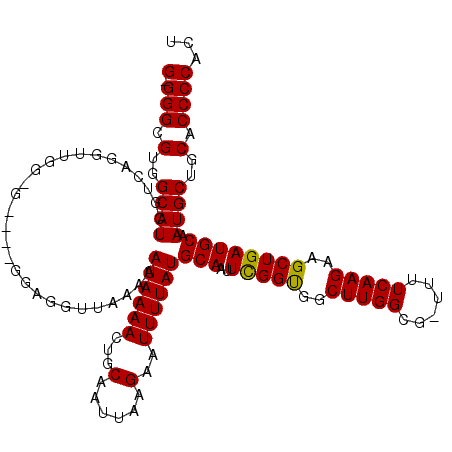
| Location | 2,807,924 – 2,808,038 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.45 |
| Mean single sequence MFE | -26.47 |
| Consensus MFE | -21.26 |
| Energy contribution | -21.50 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.930937 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2807924 114 - 22407834 AGUGGGGUGCAGCAUUGCAUCAGCUUCUUGAAAA-CGCCAAGCCACCGAAUUGCAUAAAAUUCUUAAUUGCAAUUUUAUUUUUAACCUCC----CGCCACCCUGACAAUGCCACGCCC-C ...((((((..((((((..((((...((((....-...)))).....((((((((.............))))))))..............----.......))))))))))..)))))-) ( -29.02) >DroSec_CAF1 30815 118 - 1 AGUGGGGUGCAGCAUUGCAUCAGCUUCUUGAAAA-CGCCAAGCCACCGAAUUGCAUAAAAUUCUUAAUUGCACUUUUAUUUUUAACCUCCCCCUCCCCAACCUGACGAUGCCACGCCC-C ...((((((..((((((..((((...((((....-...)))).....(((((......)))))......................................))))))))))..)))))-) ( -25.50) >DroSim_CAF1 32661 118 - 1 AGUGGGGUGCAGCAUUGCAUCAGCUUCUUGAAAA-CGCCAAGCCACCGAAUUGCAUAAAAUUCUUAAUUGCACUUUUAUUUUUAACCUCCACCUCCCCAACCUGACGAUGCCACGCCC-C ...((((((..((((((..((((...((((....-...)))).....(((((......)))))......................................))))))))))..)))))-) ( -25.50) >DroEre_CAF1 31518 107 - 1 AGUGGGGUGCAGCAUUGCAUCAGCUUCUUGAAAA-CGCCAAGCCACCGAAUUGCAUAAAAUUCUGAAUUGCAGUUUUAUUUUUAGCCC-----------ACCCAACGAUGCCACGCCC-C ...((((((..((((((.....(((.....((((-(..(((..((..(((((......)))))))..)))..)))))......)))..-----------......))))))..)))))-) ( -26.92) >DroYak_CAF1 30698 109 - 1 AGUGGGGUGCAGCAUUGCAUCAGCUACUUGAAAAUCGCCAAGCCGCCAAAUUGCAUAAAAUUCUUAAUUGCAGUUUUAUUUUUAACCC-----------ACCCCACCAUGCCACGCCCCC .((((((((.............((..((((........))))..)).((((((((.............))))))))...........)-----------))))))).............. ( -25.42) >consensus AGUGGGGUGCAGCAUUGCAUCAGCUUCUUGAAAA_CGCCAAGCCACCGAAUUGCAUAAAAUUCUUAAUUGCACUUUUAUUUUUAACCUCC____C_CCAACCUGACGAUGCCACGCCC_C ....(((((..((((((.........((((........)))).....((((((((.............)))))))).............................))))))..))))).. (-21.26 = -21.50 + 0.24)

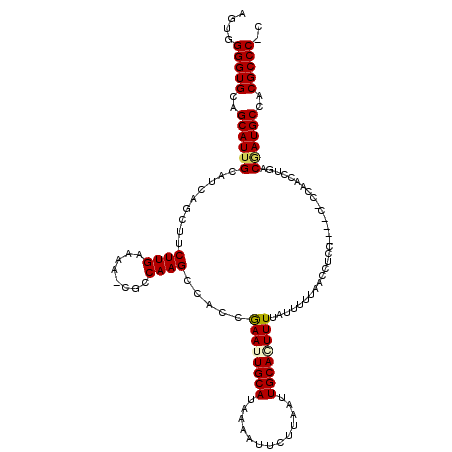
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:13 2006