| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,801,397 – 2,801,517 |
| Length | 120 |
| Max. P | 0.985592 |

| Location | 2,801,397 – 2,801,517 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.76 |
| Mean single sequence MFE | -39.30 |
| Consensus MFE | -30.18 |
| Energy contribution | -31.66 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.77 |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.985592 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
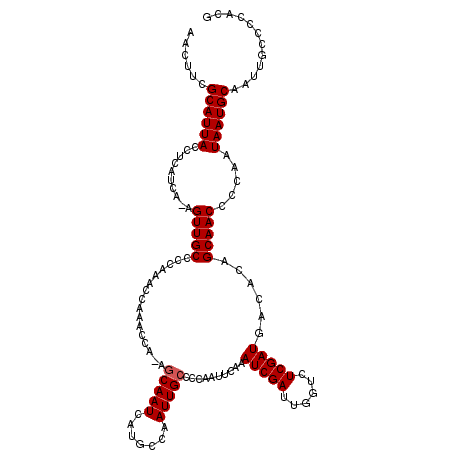
>2L_DroMel_CAF1 2801397 120 + 22407834 CGUGGGGCAAUUGCAUUAUUGGGGUUGCUGUGUCAUCGAGACUAAUCGAUUUGAAUUGGGGCAAUUGGCAUGAUUGCUUUGGUUUGGUUUGGGGCAACUUUGAUGAGGUAAUGCGAAGUU ......(((.((((.((((..((((((((..(((.....)))...((((.(..(((..((((((((.....))))))))..)))..).))))))))))))..)))).)))))))...... ( -42.80) >DroSec_CAF1 24379 119 + 1 CGUGGGGCAAUUGCAUUAUUGGGGUUGCUGCGUCAUCGAGACCAAUCGAUUUGAAUUGGGGCAAUUGGCAUGAUUGCUGUGGUUUGGUUUGGGGCAACU-UGAUGAGGUAAUGCGAAUUU (((..(((((((.((....)).)))))))((.((((((((.....((((.(..(((((.(((((((.....))))))).)))))..).)))).....))-)))))).))...)))..... ( -40.30) >DroSim_CAF1 25767 119 + 1 CGUGGGGCAAUUGCAUUAUUGGGGUUGCUGUGUCAUCGAGACCAAUCGAUUUGAAUUGGGGCAAUUGGCAUGAUUGCUGUGGUUUGGUUUGGGGCAACU-UGAUGAGGUAAUGCGAAGUU (((..(((((((.((....)).))))))).(.((((((((.....((((.(..(((((.(((((((.....))))))).)))))..).)))).....))-)))))).)....)))..... ( -37.10) >DroEre_CAF1 24801 114 + 1 CGUGGGGCAAUUGCAUUAUUGGGGUUGCUGUGUCAUCGAGACCAAUCGAUCUGAAUUGGGGCAAUUGGGAUGAUUGG-----UUCGGCUUGUGGCAACU-UGAUGAGGUAAUGCAAAGUU ..........((((((((((.(((((((..((...(((((.((((((.((((.(((((...))))).))))))))))-----)))))..))..))))))-).....)))))))))).... ( -40.00) >DroYak_CAF1 24020 114 + 1 CGUGGGGCAAUUGCAUUAUUGGGGUUGCUGUGUCAUCGACAGCAAUCGAUUUGAAUUGGGGCAAUUGUGAUGAUUGC-----UUCGCUUUGGGGCAACU-UGAUGAGGUAAUGCAAAGUU ..........((((((((((..(((((((((.......)))))))))..........(((((((((.....))))))-----)))...(..((....))-..)...)))))))))).... ( -36.30) >consensus CGUGGGGCAAUUGCAUUAUUGGGGUUGCUGUGUCAUCGAGACCAAUCGAUUUGAAUUGGGGCAAUUGGCAUGAUUGCU_UGGUUUGGUUUGGGGCAACU_UGAUGAGGUAAUGCGAAGUU ......(((.((((.((((((.(((((((.(...(((((......)))))((((((((.(((((((.....))))))).))))))))...).))))))).)))))).)))))))...... (-30.18 = -31.66 + 1.48)

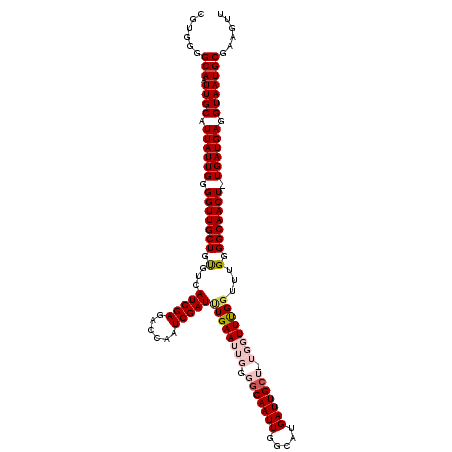
| Location | 2,801,397 – 2,801,517 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.76 |
| Mean single sequence MFE | -19.85 |
| Consensus MFE | -14.90 |
| Energy contribution | -15.10 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.838201 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2801397 120 - 22407834 AACUUCGCAUUACCUCAUCAAAGUUGCCCCAAACCAAACCAAAGCAAUCAUGCCAAUUGCCCCAAUUCAAAUCGAUUAGUCUCGAUGACACAGCAACCCCAAUAAUGCAAUUGCCCCACG ......((((((..........(((((................(((((.......)))))..........(((((......)))))......))))).....))))))............ ( -16.16) >DroSec_CAF1 24379 119 - 1 AAAUUCGCAUUACCUCAUCA-AGUUGCCCCAAACCAAACCACAGCAAUCAUGCCAAUUGCCCCAAUUCAAAUCGAUUGGUCUCGAUGACGCAGCAACCCCAAUAAUGCAAUUGCCCCACG ......((((((........-.(((((................(((((.......))))).((((((......))))))..........)))))........))))))............ ( -18.73) >DroSim_CAF1 25767 119 - 1 AACUUCGCAUUACCUCAUCA-AGUUGCCCCAAACCAAACCACAGCAAUCAUGCCAAUUGCCCCAAUUCAAAUCGAUUGGUCUCGAUGACACAGCAACCCCAAUAAUGCAAUUGCCCCACG ......((((((........-.(((((................(((((.......))))).((((((......)))))).............))))).....))))))............ ( -17.34) >DroEre_CAF1 24801 114 - 1 AACUUUGCAUUACCUCAUCA-AGUUGCCACAAGCCGAA-----CCAAUCAUCCCAAUUGCCCCAAUUCAGAUCGAUUGGUCUCGAUGACACAGCAACCCCAAUAAUGCAAUUGCCCCACG ....((((((((........-.(((((.......((((-----(((((((((..(((((...)))))..))).))))))).)))........))))).....)))))))).......... ( -24.80) >DroYak_CAF1 24020 114 - 1 AACUUUGCAUUACCUCAUCA-AGUUGCCCCAAAGCGAA-----GCAAUCAUCACAAUUGCCCCAAUUCAAAUCGAUUGCUGUCGAUGACACAGCAACCCCAAUAAUGCAAUUGCCCCACG ....((((((((........-.(((((.......((((-----((((((.....(((((...)))))......))))))).)))........))))).....)))))))).......... ( -22.20) >consensus AACUUCGCAUUACCUCAUCA_AGUUGCCCCAAACCAAACCA_AGCAAUCAUGCCAAUUGCCCCAAUUCAAAUCGAUUGGUCUCGAUGACACAGCAACCCCAAUAAUGCAAUUGCCCCACG ......((((((..........(((((................(((((.......)))))..........(((((......)))))......))))).....))))))............ (-14.90 = -15.10 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:00 2006