| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,796,358 – 2,796,528 |
| Length | 170 |
| Max. P | 0.874954 |

| Location | 2,796,358 – 2,796,456 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 77.72 |
| Mean single sequence MFE | -23.72 |
| Consensus MFE | -9.16 |
| Energy contribution | -9.88 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.39 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.843561 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
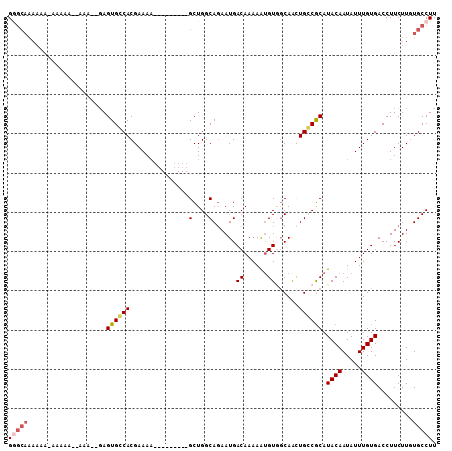
>2L_DroMel_CAF1 2796358 98 - 22407834 GGGCAAAAAAAAAAAAAAAAAC--AGUGCCACGAAAU---------GCUGGCAGAAUGACAAAAGUGUGGCAACUGGCGCAUACAAUAUUUGUGACCUUCUUGUGCCUU (((((................(--(((((((((...(---------..((.........))..).)))))).)))).((((.........)))).........))))). ( -21.90) >DroSec_CAF1 19462 77 - 1 G---------------------UGAGUGCCACGAAAA---------GCUGGCAUAAUGACAAAAAUGU--CAGUUGCCACAUACAAUAUUUGUGACCUUCUUGUGCCUU .---------------------.(((.((.((((.((---------(.(((((...(((((....)))--))..)))))..((((.....))))..))).))))))))) ( -19.80) >DroSim_CAF1 20416 100 - 1 GAGCAAAAAAAAAAAAAAAAACUGAGUGCCACGAAAA---------GCUGGCAGAAUGACAAAAAUGUGGCAACUGGCGCAUACAAUAUUUGUGACCUUCUUGUGCCUU ..........................((((((.....---------).)))))...............((((...((((((.........)))).))......)))).. ( -17.20) >DroEre_CAF1 19594 95 - 1 GGGCAAAAAACAAGUA--UAA--GAGUGGCACGAAAA---------GCUGGCAGAAUGACAAAAA-GUAGCAACUGCCGCAUACAAUAUUUGUGACCUUCUUAUGCCUU .............(((--(((--(((.((((((((..---------((.(((((....((.....-)).....)))))))........)))))).)))))))))))... ( -24.30) >DroYak_CAF1 18763 100 - 1 GGGCAAA-----AGCA--UAG--GAGUGGCACGAAAAGCUGGAAAAGCUGGCAGAAUGACAAAAAUGUGGCAAGUGCCGCAUACAAUAUUUGUGACCUUCUUAUGCCUU .......-----.(((--(((--(((.((((((((..((..(.....)..))............(((((((....)))))))......)))))).)))))))))))... ( -35.40) >consensus GGGCAAAAAA_AAAAA__AAA__GAGUGCCACGAAAA_________GCUGGCAGAAUGACAAAAAUGUGGCAACUGCCGCAUACAAUAUUUGUGACCUUCUUGUGCCUU (((((....................((((((...............(....)......((......))......)))))).((((.....)))).........))))). ( -9.16 = -9.88 + 0.72)

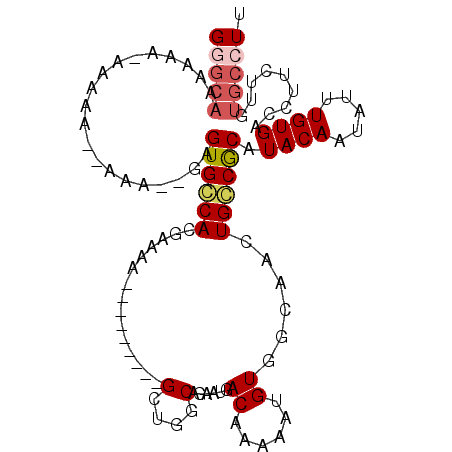
| Location | 2,796,421 – 2,796,528 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 79.82 |
| Mean single sequence MFE | -36.48 |
| Consensus MFE | -20.42 |
| Energy contribution | -21.30 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.709791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2796421 107 + 22407834 ---AUUUCGUGGCACU--GUUUUUUUUUUUUUUUUUGCCCACGGGUCAACGGGGCGUAUGGGUGUGCUGGUUAUUUUGCGGUGCCCACUUGUCUGGGCAC-CAAGAAGUGGCA ---.......(((((.--.................(((((.((......))))))).......))))).((((((((..((((((((......)))))))-)..)))))))). ( -37.81) >DroSec_CAF1 19523 89 + 1 ---UUUUCGUGGCACUCA---------------------CACGGGUCAACGGGGCGUAUGGGUGUGCUGGUUAUUUUGCGGUGCCCACUUGUCUCGGCCAAAAAGAAGUGGCA ---..((((((.......---------------------))))))....(((((((..((((..(((..........)).)..))))..)))))))((((........)))). ( -31.20) >DroEre_CAF1 19656 105 + 1 ---UUUUCGUGCCACUC--UUA--UACUUGUUUUUUGCCCACGGGUCAACGGGGCGUAUGAGUGUUCUGGUUAUUUUGUGGUGCCCACUUGUCUGGGCCC-AAAGAAGUGGCA ---......((((((((--(((--((((..(....(((((.((......))))))).)..))))).............(((.(((((......)))))))-)))).))))))) ( -40.70) >DroYak_CAF1 18832 103 + 1 AGCUUUUCGUGCCACUC--CUA--UGCU-----UUUGCCCACGGGUCAACGGGGCGUAUGAGUGUGCUGGUUAUUUUACGGUGCCCACUUGUCUGGCCCC-AAAGAAGUGGCA .........(((((((.--((.--....-----.(((((....)).))).(((((..(..((((.(((.((......)).).)).))))..)...)))))-..)).))))))) ( -36.20) >consensus ___UUUUCGUGCCACUC__UUA__U_CU_____UUUGCCCACGGGUCAACGGGGCGUAUGAGUGUGCUGGUUAUUUUGCGGUGCCCACUUGUCUGGGCCC_AAAGAAGUGGCA .........(((((((....................((((.((......))))))(.(..((((.(((.((......)).).)).))))..)).............))))))) (-20.42 = -21.30 + 0.88)

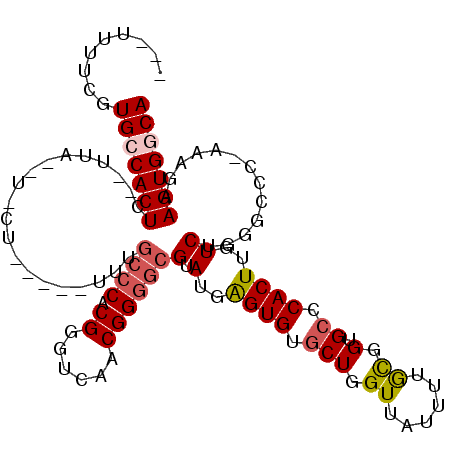
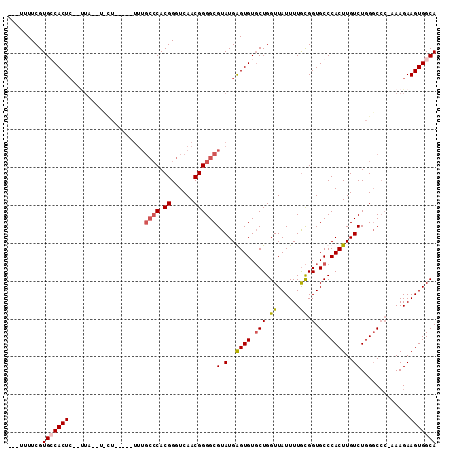
| Location | 2,796,421 – 2,796,528 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 79.82 |
| Mean single sequence MFE | -29.23 |
| Consensus MFE | -17.80 |
| Energy contribution | -19.80 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.874954 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2796421 107 - 22407834 UGCCACUUCUUG-GUGCCCAGACAAGUGGGCACCGCAAAAUAACCAGCACACCCAUACGCCCCGUUGACCCGUGGGCAAAAAAAAAAAAAAAAAC--AGUGCCACGAAAU--- ((.((((....(-(((((((......))))))))........................((((((......)).))))..................--)))).))......--- ( -29.10) >DroSec_CAF1 19523 89 - 1 UGCCACUUCUUUUUGGCCGAGACAAGUGGGCACCGCAAAAUAACCAGCACACCCAUACGCCCCGUUGACCCGUG---------------------UGAGUGCCACGAAAA--- ...(((((.((((.....)))).)))))(((((((((...((((..((..........))...)))).....))---------------------)).))))).......--- ( -19.70) >DroEre_CAF1 19656 105 - 1 UGCCACUUCUUU-GGGCCCAGACAAGUGGGCACCACAAAAUAACCAGAACACUCAUACGCCCCGUUGACCCGUGGGCAAAAAACAAGUA--UAA--GAGUGGCACGAAAA--- (((((((.((((-(((((((......))))).))).......................((((((......)).))))............--.))--))))))))......--- ( -33.50) >DroYak_CAF1 18832 103 - 1 UGCCACUUCUUU-GGGGCCAGACAAGUGGGCACCGUAAAAUAACCAGCACACUCAUACGCCCCGUUGACCCGUGGGCAAA-----AGCA--UAG--GAGUGGCACGAAAAGCU ((((((((((..-(((((......((((.((...((......))..)).)))).....))))).(((.((....))))).-----....--.))--))))))))......... ( -34.60) >consensus UGCCACUUCUUU_GGGCCCAGACAAGUGGGCACCGCAAAAUAACCAGCACACCCAUACGCCCCGUUGACCCGUGGGCAAA_____AG_A__UAA__GAGUGCCACGAAAA___ ((((((((.......(((((......)))))...........................((((((......)).))))...................))))))))......... (-17.80 = -19.80 + 2.00)

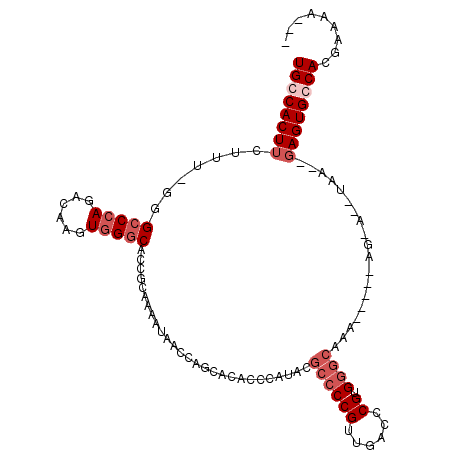
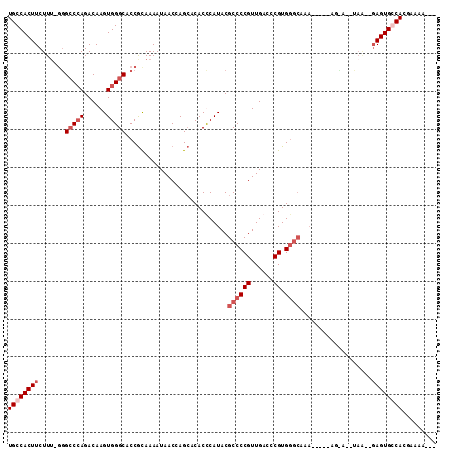
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:49:54 2006