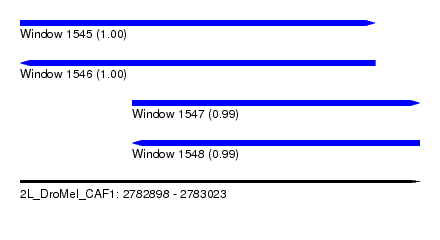
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,782,898 – 2,783,023 |
| Length | 125 |
| Max. P | 0.996601 |

| Location | 2,782,898 – 2,783,009 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 73.28 |
| Mean single sequence MFE | -43.62 |
| Consensus MFE | -34.76 |
| Energy contribution | -34.58 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.25 |
| Mean z-score | -6.25 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.68 |
| SVM RNA-class probability | 0.996320 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2782898 111 + 22407834 GGUUUCCAGAUUCCAGUGGUUAGAAACCAUCCUUAAAGGUGUCUGAAAGUAUGCAGUGAAAGGGCAAUCCACUUACAAGUUAUUGUUGCAUACUUUCAGACACCUAGGAUA ((((((..((((.....)))).))))))(((((...((((((((((((((((((((..(..((.....))(((....)))..)..))))))))))))))))))))))))). ( -47.10) >DroSec_CAF1 6141 100 + 1 GGUUUCGAGACCCCAGUGGGUAAAAAUGACCCUUUAAGGUGUCUGGAAGUAUGCAGUGAGAGGG-----------CAAUUUAUUGUUGCAUACUUUUAGACACCUAGGUUA ((((.....((((....))))......)))).....((((((((((((((((((((..(.(((.-----------...))).)..))))))))))))))))))))...... ( -41.60) >DroSim_CAF1 6250 111 + 1 GGUUUCGAGACCCCAGUGGUUAGAAAUCACCCUUUAAGGUGUCUGCAAGUAUGCAGUGAAAGGCUAAUCCACUUACAAAUUAUUGUUGCAUACUUUUAGACACCUAGGUUG ((((((..((((.....)))).))))))..(((...(((((((((.((((((((((..((((.........)))........)..)))))))))).))))))))))))... ( -38.40) >DroEre_CAF1 6104 86 + 1 GGUUUCGAGACCCCAGUGGUUAGAAAUCGCCAUUGCAGGUGCCUGAAAGUAUGCAACACAA-------------------------UGCAUACUUUCAGGCACCCUGGUCA ........((((.(((((((.(....).)))))))..((((((((((((((((((......-------------------------))))))))))))))))))..)))). ( -47.40) >consensus GGUUUCGAGACCCCAGUGGUUAGAAAUCACCCUUUAAGGUGUCUGAAAGUAUGCAGUGAAAGGG___________CAA_UUAUUGUUGCAUACUUUCAGACACCUAGGUUA ((((....))))((.(((((.....)))))......((((((((((((((((((((..(.......................)..)))))))))))))))))))).))... (-34.76 = -34.58 + -0.19)

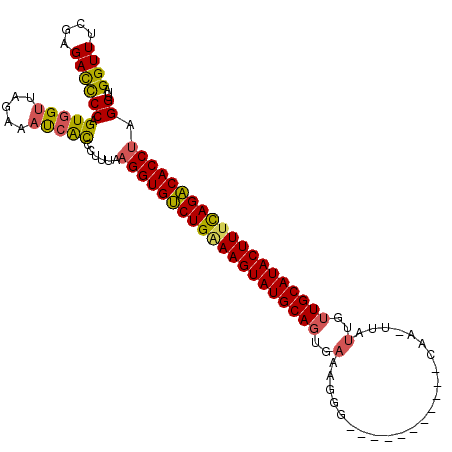
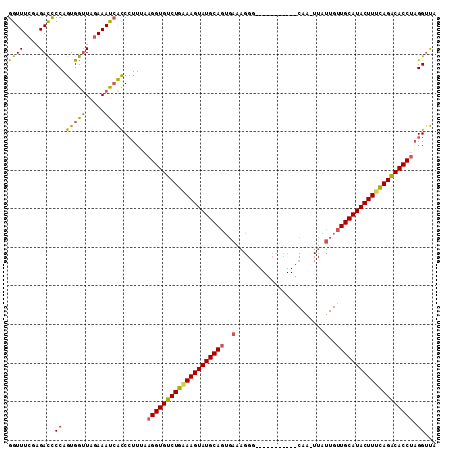
| Location | 2,782,898 – 2,783,009 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 73.28 |
| Mean single sequence MFE | -40.69 |
| Consensus MFE | -31.02 |
| Energy contribution | -31.46 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.21 |
| Mean z-score | -6.27 |
| Structure conservation index | 0.76 |
| SVM decision value | 2.72 |
| SVM RNA-class probability | 0.996601 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2782898 111 - 22407834 UAUCCUAGGUGUCUGAAAGUAUGCAACAAUAACUUGUAAGUGGAUUGCCCUUUCACUGCAUACUUUCAGACACCUUUAAGGAUGGUUUCUAACCACUGGAAUCUGGAAACC .(((((((((((((((((((((((.((((....)))).((((((.......))))))))))))))))))))))))...)))))((((((((............)))))))) ( -49.00) >DroSec_CAF1 6141 100 - 1 UAACCUAGGUGUCUAAAAGUAUGCAACAAUAAAUUG-----------CCCUCUCACUGCAUACUUCCAGACACCUUAAAGGGUCAUUUUUACCCACUGGGGUCUCGAAACC ...(((((((((((..(((((((((...........-----------.........)))))))))..))))))).....((((.......))))..))))........... ( -30.65) >DroSim_CAF1 6250 111 - 1 CAACCUAGGUGUCUAAAAGUAUGCAACAAUAAUUUGUAAGUGGAUUAGCCUUUCACUGCAUACUUGCAGACACCUUAAAGGGUGAUUUCUAACCACUGGGGUCUCGAAACC ...(((((((((((..((((((((.((((....)))).((((((.......))))))))))))))..))))))))...)))(.(((..(........)..))).)...... ( -39.70) >DroEre_CAF1 6104 86 - 1 UGACCAGGGUGCCUGAAAGUAUGCA-------------------------UUGUGUUGCAUACUUUCAGGCACCUGCAAUGGCGAUUUCUAACCACUGGGGUCUCGAAACC .((((.(((((((((((((((((((-------------------------......))))))))))))))))))).((.(((..........))).)).))))........ ( -43.40) >consensus UAACCUAGGUGUCUAAAAGUAUGCAACAAUAA_UUG___________CCCUUUCACUGCAUACUUUCAGACACCUUAAAGGGUGAUUUCUAACCACUGGGGUCUCGAAACC ...((((((((((((((((((((((...............................)))))))))))))))))))...)))..((((((........))))))........ (-31.02 = -31.46 + 0.44)

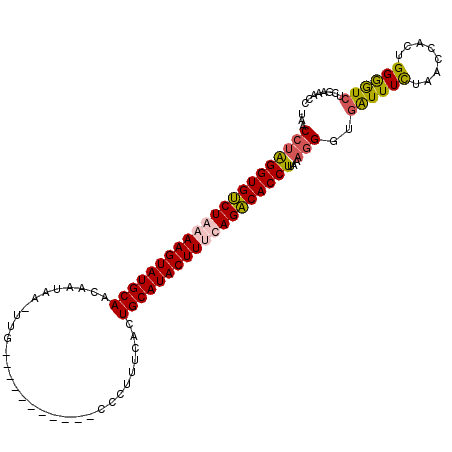
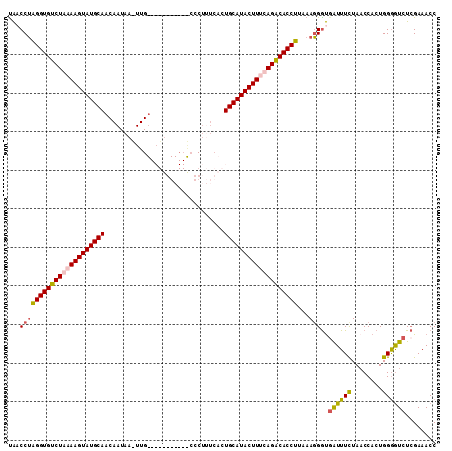
| Location | 2,782,933 – 2,783,023 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 78.54 |
| Mean single sequence MFE | -32.24 |
| Consensus MFE | -30.77 |
| Energy contribution | -30.90 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.10 |
| Mean z-score | -5.95 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.24 |
| SVM RNA-class probability | 0.990955 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2782933 90 + 22407834 AAGGUGUCUGAAAGUAUGCAGUGAAAGGGCAAUCCACUUACAAGUUAUUGUUGCAUACUUUCAGACACCUAGGAUAUUUGUUUUCUUUUC .((((((((((((((((((((..(..((.....))(((....)))..)..))))))))))))))))))))((((........)))).... ( -36.50) >DroSec_CAF1 6176 79 + 1 AAGGUGUCUGGAAGUAUGCAGUGAGAGGG-----------CAAUUUAUUGUUGCAUACUUUUAGACACCUAGGUUAUUUGUAUACUUUUC .((((((((((((((((((((..(.(((.-----------...))).)..)))))))))))))))))))).................... ( -32.70) >DroSim_CAF1 6285 85 + 1 AAGGUGUCUGCAAGUAUGCAGUGAAAGGCUAAUCCACUUACAAAUUAUUGUUGCAUACUUUUAGACACCUAGGUUGU-----UACUUUUC .(((((((((.((((((((((..((((.........)))........)..)))))))))).))))))))).......-----........ ( -27.80) >DroYak_CAF1 4874 85 + 1 AAGGUGUCUGAAAGUAUGCAGUGAAAGGGCAACCCACUUAAAAUUUAUUGUUGCAUACUUUCCGACACCUAGAUAA-----UAUCAAUGC .(((((((.((((((((((((..(..((....)).............)..)))))))))))).)))))))......-----......... ( -31.96) >consensus AAGGUGUCUGAAAGUAUGCAGUGAAAGGGCAAUCCACUUACAAUUUAUUGUUGCAUACUUUCAGACACCUAGGUUAU____UUACUUUUC .((((((((((((((((((((..(.......................)..)))))))))))))))))))).................... (-30.77 = -30.90 + 0.13)

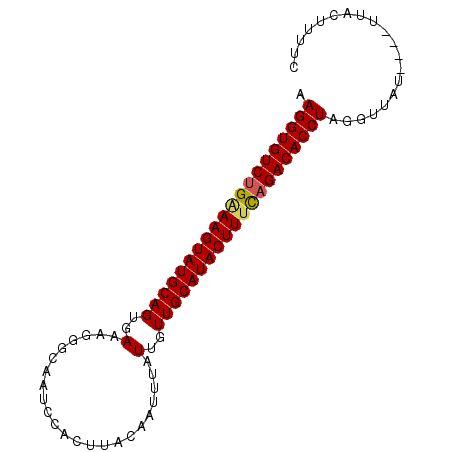
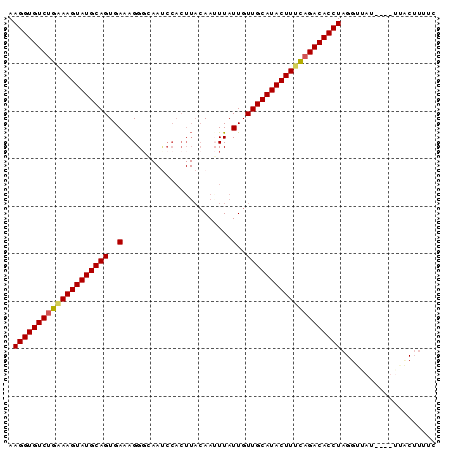
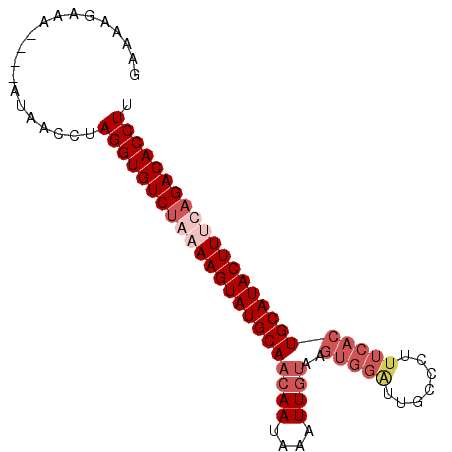
| Location | 2,782,933 – 2,783,023 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 78.54 |
| Mean single sequence MFE | -29.66 |
| Consensus MFE | -25.67 |
| Energy contribution | -28.55 |
| Covariance contribution | 2.88 |
| Combinations/Pair | 1.04 |
| Mean z-score | -6.29 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.990594 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2782933 90 - 22407834 GAAAAGAAAACAAAUAUCCUAGGUGUCUGAAAGUAUGCAACAAUAACUUGUAAGUGGAUUGCCCUUUCACUGCAUACUUUCAGACACCUU ....................((((((((((((((((((.((((....)))).((((((.......)))))))))))))))))))))))). ( -37.10) >DroSec_CAF1 6176 79 - 1 GAAAAGUAUACAAAUAACCUAGGUGUCUAAAAGUAUGCAACAAUAAAUUG-----------CCCUCUCACUGCAUACUUCCAGACACCUU ....................((((((((..(((((((((...........-----------.........)))))))))..)))))))). ( -22.35) >DroSim_CAF1 6285 85 - 1 GAAAAGUA-----ACAACCUAGGUGUCUAAAAGUAUGCAACAAUAAUUUGUAAGUGGAUUAGCCUUUCACUGCAUACUUGCAGACACCUU ........-----.......((((((((..((((((((.((((....)))).((((((.......))))))))))))))..)))))))). ( -30.80) >DroYak_CAF1 4874 85 - 1 GCAUUGAUA-----UUAUCUAGGUGUCGGAAAGUAUGCAACAAUAAAUUUUAAGUGGGUUGCCCUUUCACUGCAUACUUUCAGACACCUU .........-----......(((((((.((((((((((...((......)).((((((.......)))))))))))))))).))))))). ( -28.40) >consensus GAAAAGAAA____AUAACCUAGGUGUCUAAAAGUAUGCAACAAUAAAUUGUAAGUGGAUUGCCCUUUCACUGCAUACUUUCAGACACCUU ....................(((((((((((((((((((((((....))))..(((((.......)))))))))))))))))))))))). (-25.67 = -28.55 + 2.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:49:48 2006