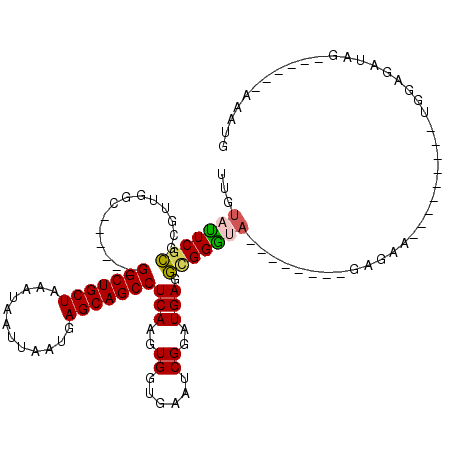
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
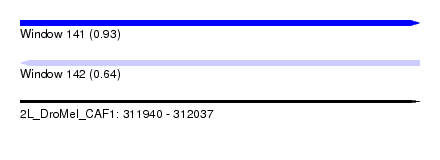
| Location | 311,940 – 312,037 |
| Length | 97 |
| Max. P | 0.928154 |

| Location | 311,940 – 312,037 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.77 |
| Mean single sequence MFE | -18.05 |
| Consensus MFE | -11.98 |
| Energy contribution | -12.48 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928154 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 311940 97 + 22407834 CAUUUUCCAUUCUAUCUUCA---------UUCUC--------UACCCGCCUCAUCCGAUUCACCACUUGAGGCUGCUCAUUAAUUAUUUAGCAGCC------GCCAACGCGAGAAUACAA ...................(---------(((((--------............................(((((((.(........).)))))))------((....)))))))).... ( -19.10) >DroSec_CAF1 16131 97 + 1 CAUUUUCCAUUCAAUCUCCA---------UUCUC--------UACCCGCCUCAUCCGAUUCACCACUUGAGGCUGCUCAUUAAUUAUUUAGCAGCC------GCCAACGCGCGAAUACAA ....................---------.....--------....(((.....................(((((((.(........).)))))))------((....)))))....... ( -16.50) >DroSim_CAF1 17951 97 + 1 CAUUUUCCAUUCUAUCUCCA---------UUCUC--------UACCCGCCUCAUCCGAUUCACCACUUGAGGCUGCUCAUUAAUUAUUUAGCAGCC------GCCAACGCGCGAAUACAA ....................---------.....--------....(((.....................(((((((.(........).)))))))------((....)))))....... ( -16.50) >DroEre_CAF1 17478 91 + 1 CAUUU------CUAUCUCCG---------UUCUC--------UACCCGCCUCAUCCGUCUCACCACUUGAGGCUGCUCAUUAAUUAUUUAGCAGCC------GCCAACGCGCGAGUACAA .....------.........---------.....--------(((.(((.....................(((((((.(........).)))))))------((....))))).)))... ( -18.40) >DroYak_CAF1 17914 99 + 1 CAUUU-------UAUCUCCGCCUCUCCUACUCUC--------UGCCCGCCUCAUCCGCUUUACCACUUGAGGCUGCUCAUUAAUUAUUUAGCAGCC------GCCAACGCGCGAAUACAA .....-------....(((((.............--------.....((.......))........(((.(((((((.(........).)))))))------..))).))).))...... ( -17.20) >DroAna_CAF1 25817 105 + 1 UAUAU------AAUUCUCCC---------UUUUCUGUUGCUGUCGCCAAUUCACCCGUUUUACCACUUGAGGCUGCUCAUUAAUUAUUUAGCAGCCGCCGCCGCCAACGCCCGACUGCAG .....------.........---------........(((.((((.........................(((((((.(........).)))))))((....)).......)))).))). ( -20.60) >consensus CAUUU______CUAUCUCCA_________UUCUC________UACCCGCCUCAUCCGAUUCACCACUUGAGGCUGCUCAUUAAUUAUUUAGCAGCC______GCCAACGCGCGAAUACAA ..............................................(((.....................(((((((.(........).)))))))......((....)))))....... (-11.98 = -12.48 + 0.50)

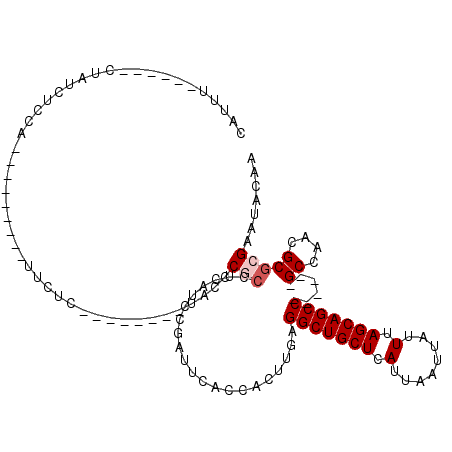
| Location | 311,940 – 312,037 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.77 |
| Mean single sequence MFE | -24.72 |
| Consensus MFE | -17.61 |
| Energy contribution | -17.50 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.635941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 311940 97 - 22407834 UUGUAUUCUCGCGUUGGC------GGCUGCUAAAUAAUUAAUGAGCAGCCUCAAGUGGUGAAUCGGAUGAGGCGGGUA--------GAGAA---------UGAAGAUAGAAUGGAAAAUG ...(((((((((.((((.------(((((((............))))))))))))).....(((.(......).))).--------)))))---------)).................. ( -22.80) >DroSec_CAF1 16131 97 - 1 UUGUAUUCGCGCGUUGGC------GGCUGCUAAAUAAUUAAUGAGCAGCCUCAAGUGGUGAAUCGGAUGAGGCGGGUA--------GAGAA---------UGGAGAUUGAAUGGAAAAUG ((.(((((((.((((.(.------(((((((............)))))))(((.....)))..).))))..)))))))--------.))..---------.................... ( -23.40) >DroSim_CAF1 17951 97 - 1 UUGUAUUCGCGCGUUGGC------GGCUGCUAAAUAAUUAAUGAGCAGCCUCAAGUGGUGAAUCGGAUGAGGCGGGUA--------GAGAA---------UGGAGAUAGAAUGGAAAAUG ((.(((((((.((((.(.------(((((((............)))))))(((.....)))..).))))..)))))))--------.))..---------.................... ( -23.40) >DroEre_CAF1 17478 91 - 1 UUGUACUCGCGCGUUGGC------GGCUGCUAAAUAAUUAAUGAGCAGCCUCAAGUGGUGAGACGGAUGAGGCGGGUA--------GAGAA---------CGGAGAUAG------AAAUG ((.(((((((.((((.((------(((((((............)))))))(((.....)))).).))))..)))))))--------.))..---------.........------..... ( -26.80) >DroYak_CAF1 17914 99 - 1 UUGUAUUCGCGCGUUGGC------GGCUGCUAAAUAAUUAAUGAGCAGCCUCAAGUGGUAAAGCGGAUGAGGCGGGCA--------GAGAGUAGGAGAGGCGGAGAUA-------AAAUG ((((.(((.(((.((.((------..(((((................((((((.((......))...)))))).))))--------)...))....)).)))))))))-------).... ( -25.63) >DroAna_CAF1 25817 105 - 1 CUGCAGUCGGGCGUUGGCGGCGGCGGCUGCUAAAUAAUUAAUGAGCAGCCUCAAGUGGUAAAACGGGUGAAUUGGCGACAGCAACAGAAAA---------GGGAGAAUU------AUAUA .(((.(((((.(((.....))).)(((((((............)))))))(((..((......))..))).....)))).)))........---------.........------..... ( -26.30) >consensus UUGUAUUCGCGCGUUGGC______GGCUGCUAAAUAAUUAAUGAGCAGCCUCAAGUGGUGAAUCGGAUGAGGCGGGUA________GAGAA_________UGGAGAUAG______AAAUG ...(((((((..............(((((((............)))))))(((..((......))..))).))))))).......................................... (-17.61 = -17.50 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:25:37 2006