
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,730,700 – 2,730,790 |
| Length | 90 |
| Max. P | 0.657658 |

| Location | 2,730,700 – 2,730,790 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 83.51 |
| Mean single sequence MFE | -27.34 |
| Consensus MFE | -16.30 |
| Energy contribution | -16.42 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.580381 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
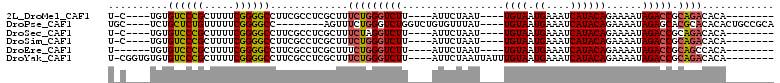
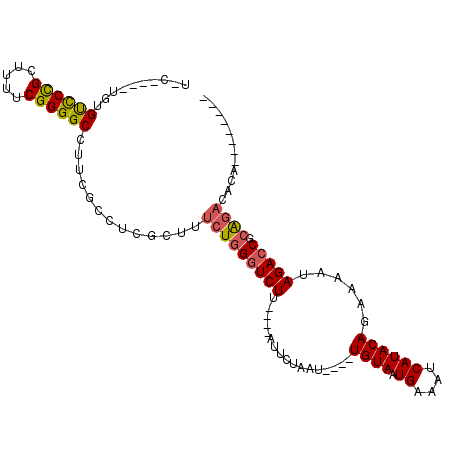
>2L_DroMel_CAF1 2730700 90 + 22407834 U-C----UGUGUCCCGCUUUUCGGGGCCUUCGCCUCGCUUUCUGGGUCUU----AUUCUAAU----UGUAAUGAAAUCAUACAGAAAAUAGACCGCAGACACA-------- .-.----.(((((..((....((((((....)))))).......(((((.----.((((...----.(((.((....)))))))))...))))))).))))).-------- ( -27.10) >DroPse_CAF1 185046 95 + 1 UGC----UCUGCUCUGUUUUUCGGGGCC--------AGUUUCUGGGUCUGGUCUGUGUUUAU----UGUAAUGAAAUCAUACAGAAAAUAGAGCACGCACACACUGCCGCA .((----..((((((((((.(((((.((--------((...)))).)))))(((((((..((----(.......))).))))))))))))))))).(((.....))).)). ( -30.00) >DroSec_CAF1 149506 90 + 1 U-C----UGUGUCCCGCUUUUCGGGGCCUUCGCCUCGCUUUCUAGGUCUU----AUUCUAAU----UGUAAUGAAAUCAUACAGAAAAUAGACCGCAGACACA-------- .-.----.(((((..((....((((((....)))))).......(((((.----.((((...----.(((.((....)))))))))...))))))).))))).-------- ( -27.30) >DroSim_CAF1 153589 90 + 1 U-C----UGUGUCCCGCUUUUCGGGGCCUUCGCCUCGCUUUCUGGGUCUU----AUUCUAAU----UGUAAUGAAAUCAUACAGAAAAUAGACCGCAGACACA-------- .-.----.(((((..((....((((((....)))))).......(((((.----.((((...----.(((.((....)))))))))...))))))).))))).-------- ( -27.10) >DroEre_CAF1 167827 89 + 1 U------UGUGUCCCGCUUUUCGGGGCCUUCGCCUCGCUUUCUGGGUCUU----AUUCUAAU----UGUAAUGAAAUCAUACAGAAAAUAGACCGCAGCCACA-------- .------((((..........((((((....))))))....((((((((.----.((((...----.(((.((....)))))))))...))))).))).))))-------- ( -22.80) >DroYak_CAF1 158564 98 + 1 U-CGGUGUGUGUCCCGCUUUUCGGGGCCUUCGCCUCGCUUUCUGGGUCUU----AUUCUAAUUAUUUGUAAUGAAAUCAUACAGAAAAUAGACCGCAGACACA-------- .-.((((...((((((.....))))))...))))......(((((((((.----......(((.((((((.((....)))))))).)))))))).))))....-------- ( -29.71) >consensus U_C____UGUGUCCCGCUUUUCGGGGCCUUCGCCUCGCUUUCUGGGUCUU____AUUCUAAU____UGUAAUGAAAUCAUACAGAAAAUAGACCGCAGACACA________ ..........((((((.....)))))).............(((((((((.................((((.((....))))))......))))).))))............ (-16.30 = -16.42 + 0.11)
| Location | 2,730,700 – 2,730,790 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 83.51 |
| Mean single sequence MFE | -28.25 |
| Consensus MFE | -18.40 |
| Energy contribution | -19.07 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.657658 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
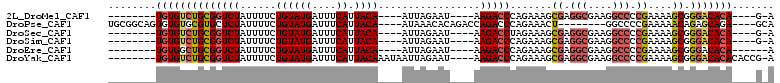
>2L_DroMel_CAF1 2730700 90 - 22407834 --------UGUGUCUGCGGUCUAUUUUCUGUAUGAUUUCAUUACA----AUUAGAAU----AAGACCCAGAAAGCGAGGCGAAGGCCCCGAAAAGCGGGACACA----G-A --------(((((((((((((((((((.((((((....)).))))----...)))))----.))))).......((.(((....))).))....)).)))))))----.-. ( -30.20) >DroPse_CAF1 185046 95 - 1 UGCGGCAGUGUGUGCGUGCUCUAUUUUCUGUAUGAUUUCAUUACA----AUAAACACAGACCAGACCCAGAAACU--------GGCCCCGAAAAACAGAGCAGA----GCA .((.(((.....))).((((((.(((((((((((....)).))))----...........((((.........))--------))....)))))..))))))..----)). ( -20.10) >DroSec_CAF1 149506 90 - 1 --------UGUGUCUGCGGUCUAUUUUCUGUAUGAUUUCAUUACA----AUUAGAAU----AAGACCUAGAAAGCGAGGCGAAGGCCCCGAAAAGCGGGACACA----G-A --------(((((((((((((((((((.((((((....)).))))----...)))))----.))))).......((.(((....))).))....)).)))))))----.-. ( -30.60) >DroSim_CAF1 153589 90 - 1 --------UGUGUCUGCGGUCUAUUUUCUGUAUGAUUUCAUUACA----AUUAGAAU----AAGACCCAGAAAGCGAGGCGAAGGCCCCGAAAAGCGGGACACA----G-A --------(((((((((((((((((((.((((((....)).))))----...)))))----.))))).......((.(((....))).))....)).)))))))----.-. ( -30.20) >DroEre_CAF1 167827 89 - 1 --------UGUGGCUGCGGUCUAUUUUCUGUAUGAUUUCAUUACA----AUUAGAAU----AAGACCCAGAAAGCGAGGCGAAGGCCCCGAAAAGCGGGACACA------A --------((((.((((((((((((((.((((((....)).))))----...)))))----.))))).......((.(((....))).))....))))..))))------. ( -28.60) >DroYak_CAF1 158564 98 - 1 --------UGUGUCUGCGGUCUAUUUUCUGUAUGAUUUCAUUACAAAUAAUUAGAAU----AAGACCCAGAAAGCGAGGCGAAGGCCCCGAAAAGCGGGACACACACCG-A --------((((((((((((((...(((((....((((......))))...))))).----.))))).......((.(((....))).))....)).))))))).....-. ( -29.80) >consensus ________UGUGUCUGCGGUCUAUUUUCUGUAUGAUUUCAUUACA____AUUAGAAU____AAGACCCAGAAAGCGAGGCGAAGGCCCCGAAAAGCGGGACACA____G_A ........((((((((((((((......((((((....)).)))).................))))).......((.(((....))).))....)).)))))))....... (-18.40 = -19.07 + 0.67)
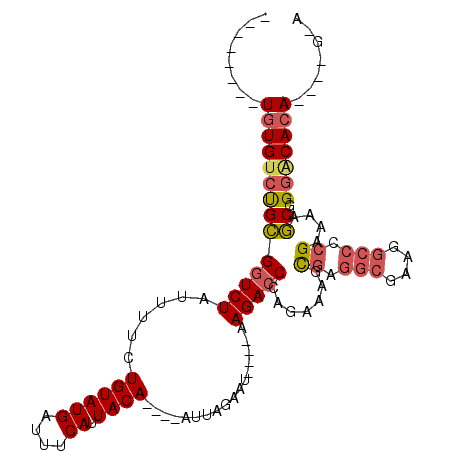
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:49:32 2006