| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,721,874 – 2,721,980 |
| Length | 106 |
| Max. P | 0.994053 |

| Location | 2,721,874 – 2,721,980 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 75.51 |
| Mean single sequence MFE | -22.18 |
| Consensus MFE | -8.87 |
| Energy contribution | -9.07 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.40 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.828696 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2721874 106 + 22407834 -------GA-CAAAAAAGGAUGACACACAAUAGUUGGGACAUUGUUUUUGACAUGACUUUCUAUCAAAAACAAAUUUGCCCUUCUUUGUGCUUCUAUUAUUUU----GUAUUUC-CCAU -------.(-(((((.((...((..(((((.((..(((.((((((((((((...(.....)..))))))))))...)))))..)))))))..))..)).))))----)).....-.... ( -23.60) >DroVir_CAF1 204429 99 + 1 -------AA-AAGAAAAGUAUGA----CAAUAGUUGGGCUAUUGUUUUUGACAUGACUUUCUAUCAAAAACAAAUUUGGUCUUCUUUGU---UGUGUUACUUU----GUGU-CGUCUUU -------.(-((((((((((..(----((((((..((((((((((((((((...(.....)..))))))))))...))))))...))))---)))..))))))----....-..))))) ( -24.50) >DroPse_CAF1 170055 106 + 1 -------GA-AAAAAAACGAUGACACACAAUAGUUGGGACAUUGUUUUUGACAUGACUUUCUAUCAAAAACAAAUUUGCCCUUCUUUGUUCUCCUAUUAUUUCUAUCCUAUU-----AU -------((-((......((.((...((((.((..(((.((((((((((((...(.....)..))))))))))...)))))..))))))))))......)))).........-----.. ( -16.30) >DroWil_CAF1 252912 110 + 1 AUAACGACAGGAAAAAAGGAUGACAUACAAUAGUUUGGACAUUGUUUUUGACAUGACUUUCUAUCAAAAACAAAUUUGUCCUUCUUU-UGUACACAUUAUUUU----GUGCUCU-U--- .....((((.((((....((((...(((((.((...(((((((((((((((...(.....)..))))))))))...)))))..)).)-))))..)))).))))----.)).)).-.--- ( -25.30) >DroMoj_CAF1 208537 99 + 1 -------AG-AAGUAAAGUAUGA----CAAUAGUUGGGCUAUUGUUUUUGACAUGACUUUCUAUCAAAAACAAAUUUGUCCUUCAUUGU---UGCAAUACUUU----GUGUUCGUCU-A -------.(-(((((((((((..----(((((((.((((..((((((((((...(.....)..))))))))))....))))...)))))---))..)))))))----)).)))....-. ( -25.90) >DroAna_CAF1 239012 113 + 1 AUAAAAACA-UAAAAAAGGAUGACAUACAAUAGUUGGGACAUUGUUUCUGACAUGACUUUCUAUCAAAAACAAAUUUGCCCUUCUUUGUUCUCCUAUUAUUUU----GUACUGU-CUAU ........(-(((((.((((.((((......((..(((.((((((((.(((...(.....)..))).))))))...)))))..)).)))).))))....))))----)).....-.... ( -17.50) >consensus _______AA_AAAAAAAGGAUGACA_ACAAUAGUUGGGACAUUGUUUUUGACAUGACUUUCUAUCAAAAACAAAUUUGCCCUUCUUUGU_CUCCUAUUAUUUU____GUAUUCG_CUAU ...................................(((...((((((((((...(.....)..)))))))))).....)))...................................... ( -8.87 = -9.07 + 0.19)


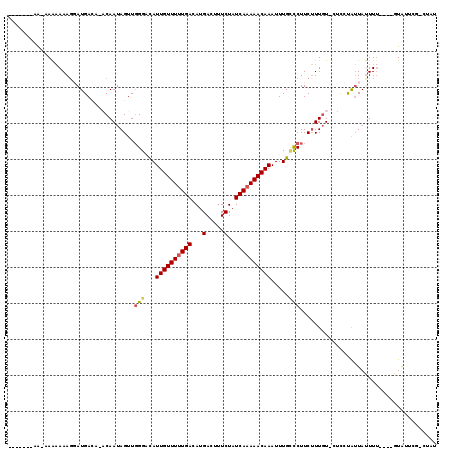
| Location | 2,721,874 – 2,721,980 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 75.51 |
| Mean single sequence MFE | -23.47 |
| Consensus MFE | -14.86 |
| Energy contribution | -15.08 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.63 |
| SVM decision value | 2.45 |
| SVM RNA-class probability | 0.994053 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2721874 106 - 22407834 AUGG-GAAAUAC----AAAAUAAUAGAAGCACAAAGAAGGGCAAAUUUGUUUUUGAUAGAAAGUCAUGUCAAAAACAAUGUCCCAACUAUUGUGUGUCAUCCUUUUUUG-UC------- ....-.....((----((((.....((.((((((((..(((((...((((((((((((........)))))))))))))).)))..)).)))))).))......)))))-).------- ( -28.20) >DroVir_CAF1 204429 99 - 1 AAAGACG-ACAC----AAAGUAACACA---ACAAAGAAGACCAAAUUUGUUUUUGAUAGAAAGUCAUGUCAAAAACAAUAGCCCAACUAUUG----UCAUACUUUUCUU-UU------- (((((..-....----((((((..(((---(...............((((((((((((........))))))))))))(((.....))))))----)..))))))))))-).------- ( -16.20) >DroPse_CAF1 170055 106 - 1 AU-----AAUAGGAUAGAAAUAAUAGGAGAACAAAGAAGGGCAAAUUUGUUUUUGAUAGAAAGUCAUGUCAAAAACAAUGUCCCAACUAUUGUGUGUCAUCGUUUUUUU-UC------- ..-----....(((.(((((..((.((...((((((..(((((...((((((((((((........)))))))))))))).)))..)).))))...))))..))))).)-))------- ( -24.20) >DroWil_CAF1 252912 110 - 1 ---A-AGAGCAC----AAAAUAAUGUGUACA-AAAGAAGGACAAAUUUGUUUUUGAUAGAAAGUCAUGUCAAAAACAAUGUCCAAACUAUUGUAUGUCAUCCUUUUUUCCUGUCGUUAU ---.-..(((((----((((..(((((((((-(.((..(((((...((((((((((((........)))))))))))))))))...)).))))))).)))..))).....))).))).. ( -25.70) >DroMoj_CAF1 208537 99 - 1 U-AGACGAACAC----AAAGUAUUGCA---ACAAUGAAGGACAAAUUUGUUUUUGAUAGAAAGUCAUGUCAAAAACAAUAGCCCAACUAUUG----UCAUACUUUACUU-CU------- .-....(((...----(((((((.(((---(.......((......((((((((((((........))))))))))))...))......)))----).)))))))..))-).------- ( -21.12) >DroAna_CAF1 239012 113 - 1 AUAG-ACAGUAC----AAAAUAAUAGGAGAACAAAGAAGGGCAAAUUUGUUUUUGAUAGAAAGUCAUGUCAGAAACAAUGUCCCAACUAUUGUAUGUCAUCCUUUUUUA-UGUUUUUAU ..((-(((....----((((....((((((((((((..(((((...((((((((((((........)))))))))))))).)))..)).))))...)).)))).)))).-))))).... ( -25.40) >consensus AUAG_AGAACAC____AAAAUAAUAGAAG_ACAAAGAAGGGCAAAUUUGUUUUUGAUAGAAAGUCAUGUCAAAAACAAUGUCCCAACUAUUGU_UGUCAUCCUUUUUUU_UC_______ ......................................(((((...((((((((((((........))))))))))))))))).................................... (-14.86 = -15.08 + 0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:49:23 2006