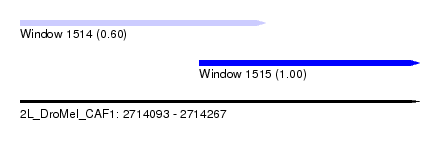
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,714,093 – 2,714,267 |
| Length | 174 |
| Max. P | 0.999346 |

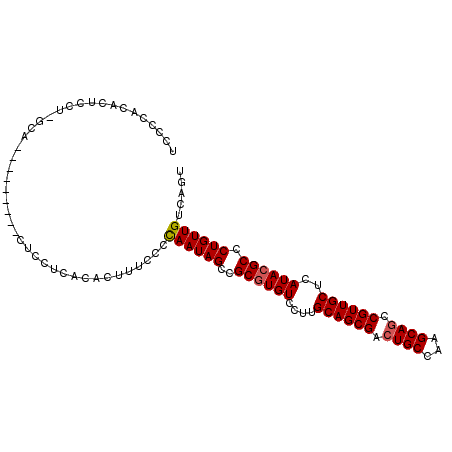
| Location | 2,714,093 – 2,714,200 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 84.28 |
| Mean single sequence MFE | -23.18 |
| Consensus MFE | -18.80 |
| Energy contribution | -18.80 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.600635 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2714093 107 + 22407834 UGUGUUCUGACUGCUCCACUCCUUUUGAAUAUGCAGAACGUCACUUCCACUGCCGGCUGCUCCUCGAGCUGCUCGCACUCACCACACUCCUUGCA---------CUUCUCACACUU .((((..(((((((..................))))..............(((.(((.((((...)))).))).))).)))..))))........---------............ ( -21.07) >DroSec_CAF1 133021 116 + 1 UGUGAGCUGACUGCUCCACUCCUUUUGAAUAUGCAGAACGUCACUUCCACUGCCGGCCGCUCCUCGAGCUGCUCGCACUCCCCACACUCCUGGCACUCCUUUCACUCCUCACACUU ((((((.(((((((..................))))..............(((.(((.((((...)))).))).)))....(((......)))........)))...))))))... ( -23.97) >DroSim_CAF1 136464 116 + 1 UGUGAGCUGACUGCGCCACUCCUUUUGAAUAUGCAGAACGUCACUUCCACUGCCGGCCGCUCCUCGAGCUGCUCGCACUCCCCACACUCCUUGCACUCCUUUCACUCCUCACACUU ((((((.((((((((....((.....))...)))))..............(((.(((.((((...)))).))).)))........................)))...))))))... ( -23.50) >DroEre_CAF1 151171 97 + 1 UGUGAGUUGACUGCUCCACUCCUUUUGAAUAUGCAGGACGUCAUUUCCACUGCCGGCCGCUCCUCGAGCUGCUCGCACUCCUCACAG------------------UUCACAC-CCU ((((((..(.((((..................))))..............(((.(((.((((...)))).))).))))..)))))).------------------.......-... ( -24.17) >consensus UGUGAGCUGACUGCUCCACUCCUUUUGAAUAUGCAGAACGUCACUUCCACUGCCGGCCGCUCCUCGAGCUGCUCGCACUCCCCACACUCCUUGCA_________CUCCUCACACUU ((((......((((..................))))..............(((.(((.((((...)))).))).))).....)))).............................. (-18.80 = -18.80 + -0.00)

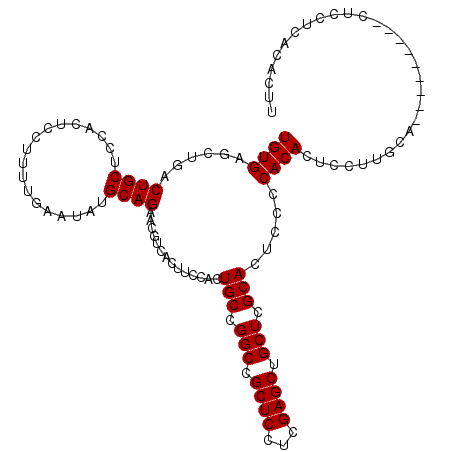
| Location | 2,714,171 – 2,714,267 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 75.42 |
| Mean single sequence MFE | -25.22 |
| Consensus MFE | -20.81 |
| Energy contribution | -21.30 |
| Covariance contribution | 0.49 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.39 |
| Structure conservation index | 0.83 |
| SVM decision value | 3.53 |
| SVM RNA-class probability | 0.999346 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2714171 96 + 22407834 UCACCACACUCCUUGCA---------CUUCUCACACUUUCCCCAAUAGGCGCAUGUCCUUGCAGCGAAUGCCAAGCAGCCGUUGCUCAUACGCCCUGUUGUCAGU .................---------................(((((((((.(((.....((((((..(((...)))..)))))).))).)).)))))))..... ( -23.40) >DroSec_CAF1 133099 105 + 1 UCCCCACACUCCUGGCACUCCUUUCACUCCUCACACUUUCCCCAAUAGCCGCGUGUCCUUGCAGCGACUGCCAAGCAGUCGUUGCUCAUACGCCCUGUUGUCAGU ...(((......)))...........................((((((..((((((....(((((((((((...)))))))))))..)))))).))))))..... ( -31.70) >DroSim_CAF1 136542 105 + 1 UCCCCACACUCCUUGCACUCCUUUCACUCCUCACACUUUCCCUAAUAGCCGCGUGUCCUUGCAGCGACUGCCAAGCAGUCGUUGCUCAUACGCCCUGUUGUCAGU ..........................................((((((..((((((....(((((((((((...)))))))))))..)))))).))))))..... ( -29.00) >DroEre_CAF1 151249 74 + 1 UCCUCACAG------------------UUCACAC-CCUUUCCCAAUAGCCGCGUGUCCUUGCAGC------CAA------GUUGCUCAUACGCCCUGUUGUCACU .........------------------.......-.......((((((..((((((....(((((------...------)))))..)))))).))))))..... ( -16.20) >DroYak_CAF1 140578 78 + 1 UCCUCACA---------------------------CUUUUCCCAAUAGCCGCGUGUCCUUGCAGCGACUGCCAAGCAGCCGUUGCUCAUACGCCCUGUUGUCAGU ........---------------------------.......((((((..((((((....((((((.((((...)))).))))))..)))))).))))))..... ( -25.80) >consensus UCCCCACACUCCU_GCA_________CUCCUCACACUUUCCCCAAUAGCCGCGUGUCCUUGCAGCGACUGCCAAGCAGCCGUUGCUCAUACGCCCUGUUGUCAGU ..........................................((((((..((((((....((((((.((((...)))).))))))..)))))).))))))..... (-20.81 = -21.30 + 0.49)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:49:16 2006