
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,693,973 – 2,694,070 |
| Length | 97 |
| Max. P | 0.861849 |

| Location | 2,693,973 – 2,694,070 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 79.91 |
| Mean single sequence MFE | -28.58 |
| Consensus MFE | -22.60 |
| Energy contribution | -22.52 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.861849 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
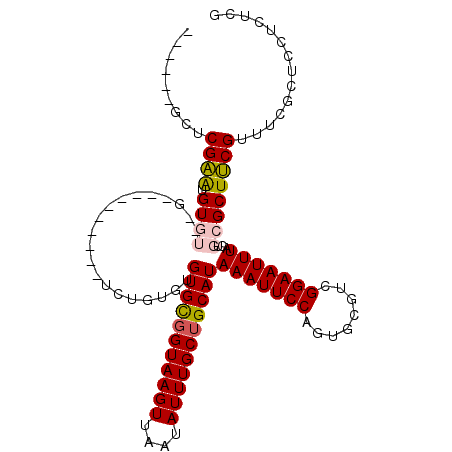
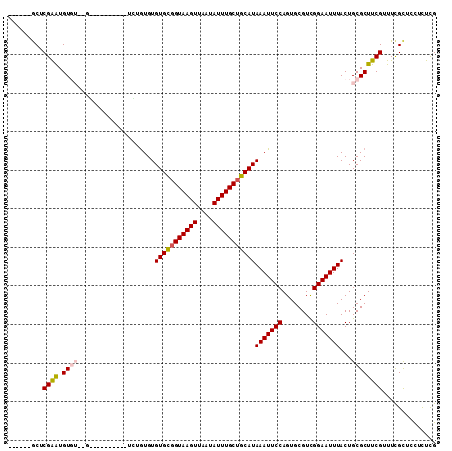
>2L_DroMel_CAF1 2693973 97 + 22407834 ------GCUCGAAUGUGUUUGU--G------UGUGUGUGUGUGGUAAGUUAAUAUUUGCUGCAUAAAUUCCAGUGCGUCGGAAUUUACUGCGCUUCGUUUUGCUCUUCUCG ------((.((((.((((..((--(------......((((..((((((....))))))..))))((((((........))))))))).))))))))....))........ ( -25.40) >DroPse_CAF1 141755 105 + 1 CCCUGUGCACGGGUGUGUACGGAUA------UCUGUGUGUGCGGUAAGUUAAUAUUUGCUGCAUAAAUUCCAGUGCGUUGGAAUUUACUGCGCUCCGUUUCGUUCCUUUUG ..((((((((....))))))))...------...(((((((((((((((....)))))))))))(((((((((....)))))))))...)))).................. ( -33.60) >DroEre_CAF1 128123 91 + 1 ------GCUCGAAUGU--------------GUGUGUGUGUGUGGUAAGUUAAUAUUUGCUGCAUAAAUUCCAGUGCGUCGGAAUUUACUGCGCUUCGUUUCGCCGCUCUCG ------((.((((.((--------------(((.(((((((..((((((....))))))..))))((((((........))))))))))))))))))....))........ ( -24.30) >DroYak_CAF1 118190 105 + 1 ------GCUCGAAUGUGUUAGUAAGUCUGUGUAUGUGUGUGUUGUAAGUUAAUAUUUGCUGCAUAAAUUCCAGUGCGUCGGAAUUUACUGCGCUUCGUUUCGCUCCUCUCG ------((.((((.((((.(((((((((((((((...(((((.((((((....)))))).))))).......))))).))).)))))))))))))))....))........ ( -29.00) >DroAna_CAF1 196540 88 + 1 -----UACCCGAAUGU------------------AUGUGUGCGGUAAGUUAAUAUUUGCUGCAUAAAUUCCAGUGCGUUGGAAUUUACUGCGCUUCGUUUCGCUCUUUUCG -----....((((...------------------..(((((((((((((....))))))))).((((((((((....))))))))))..)))).....))))......... ( -25.60) >DroPer_CAF1 138636 105 + 1 CCCUGUGCACGGGUGUGUACGGAUA------UCUGUGUGUGCGGUAAGUUAAUAUUUGCUGCAUAAAUUCCAGUGCGUUGGAAUUUACUGCGCUCCGUUUCGUUCCUUUUG ..((((((((....))))))))...------...(((((((((((((((....)))))))))))(((((((((....)))))))))...)))).................. ( -33.60) >consensus ______GCUCGAAUGUGU__G__________UCUGUGUGUGCGGUAAGUUAAUAUUUGCUGCAUAAAUUCCAGUGCGUCGGAAUUUACUGCGCUUCGUUUCGCUCCUCUCG .........((((.((((....................(((((((((((....)))))))))))(((((((........)))))))...)))))))).............. (-22.60 = -22.52 + -0.08)

| Location | 2,693,973 – 2,694,070 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 79.91 |
| Mean single sequence MFE | -18.87 |
| Consensus MFE | -15.11 |
| Energy contribution | -14.42 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.11 |
| Mean z-score | -0.99 |
| Structure conservation index | 0.80 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.514745 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
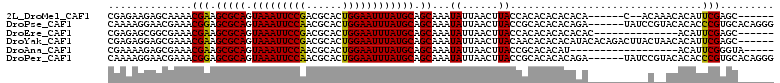
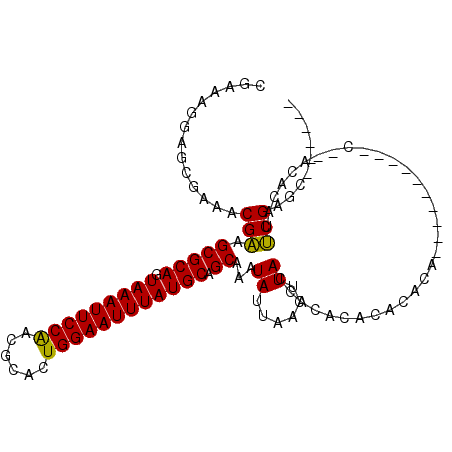
>2L_DroMel_CAF1 2693973 97 - 22407834 CGAGAAGAGCAAAACGAAGCGCAGUAAAUUCCGACGCACUGGAAUUUAUGCAGCAAAUAUUAACUUACCACACACACACA------C--ACAAACACAUUCGAGC------ ((((....((........))(((.(((((((((......)))))))))))).............................------.--.........))))...------ ( -15.80) >DroPse_CAF1 141755 105 - 1 CAAAAGGAACGAAACGGAGCGCAGUAAAUUCCAACGCACUGGAAUUUAUGCAGCAAAUAUUAACUUACCGCACACACAGA------UAUCCGUACACACCCGUGCACAGGG .............(((((.((((.(((((((((......)))))))))))).((...............)).......).------..))))).....(((.......))) ( -22.96) >DroEre_CAF1 128123 91 - 1 CGAGAGCGGCGAAACGAAGCGCAGUAAAUUCCGACGCACUGGAAUUUAUGCAGCAAAUAUUAACUUACCACACACACACAC--------------ACAUUCGAGC------ ((((..((......))..(((((.(((((((((......)))))))))))).))...........................--------------...))))...------ ( -17.20) >DroYak_CAF1 118190 105 - 1 CGAGAGGAGCGAAACGAAGCGCAGUAAAUUCCGACGCACUGGAAUUUAUGCAGCAAAUAUUAACUUACAACACACACAUACACAGACUUACUAACACAUUCGAGC------ ((((.(..((........))(((.(((((((((......)))))))))))).............................................).))))...------ ( -16.60) >DroAna_CAF1 196540 88 - 1 CGAAAAGAGCGAAACGAAGCGCAGUAAAUUCCAACGCACUGGAAUUUAUGCAGCAAAUAUUAACUUACCGCACACAU------------------ACAUUCGGGUA----- ((((....((........))(((.(((((((((......))))))))))))..........................------------------...))))....----- ( -17.70) >DroPer_CAF1 138636 105 - 1 CAAAAGGAACGAAACGGAGCGCAGUAAAUUCCAACGCACUGGAAUUUAUGCAGCAAAUAUUAACUUACCGCACACACAGA------UAUCCGUACACACCCGUGCACAGGG .............(((((.((((.(((((((((......)))))))))))).((...............)).......).------..))))).....(((.......))) ( -22.96) >consensus CGAAAGGAGCGAAACGAAGCGCAGUAAAUUCCAACGCACUGGAAUUUAUGCAGCAAAUAUUAACUUACCACACACACACA__________C__ACACAUUCGAGC______ ..............(((.(((((.(((((((((......)))))))))))).))...((......))................................)))......... (-15.11 = -14.42 + -0.69)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:49:13 2006