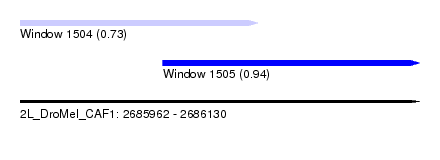
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,685,962 – 2,686,130 |
| Length | 168 |
| Max. P | 0.938335 |

| Location | 2,685,962 – 2,686,062 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 76.87 |
| Mean single sequence MFE | -27.60 |
| Consensus MFE | -13.35 |
| Energy contribution | -13.72 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.725582 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2685962 100 + 22407834 UUUUUUUCAUCCU--GC---CCUUCCAUGGGCUUCCUGGCCUGUGGAGCCGCGUGUCUUGCUCAUUGCCAGGCAUUCGCUCAUUUUCAAAUCGGAUUUUAUCAAC ........((((.--((---..(((((((((((....)))))))))))..))((((((.((.....)).)))))).................))))......... ( -30.60) >DroVir_CAF1 160587 91 + 1 CUUUUGCCACCAUUC-------UUCGUUGGGGG-------AGUCCGUGCCGCGUGCCUUGCUCAUUGCCAGGCAUUCGCUCAUUUUCAAAUCGGAUUUUAUCAAC ...............-------...(((((..(-------((((((.((.(.((((((.((.....)).)))))).)))............)))))))..))))) ( -25.42) >DroPse_CAF1 132835 93 + 1 UUUGUUUCGUCCU--GGGGGCGGUCCUUGAGC----------CUUGAGCCGCGUGUCUUGCUCAUUGCCAGGCAUUCGCUCAUUUUCAAAUCGGAUUUUAUCAAC .......(((((.--..)))))(((((((((.----------..(((((.(.((((((.((.....)).)))))).))))))..)))))...))))......... ( -27.60) >DroEre_CAF1 120156 99 + 1 UUUUUUUCAUCCU--GU---UCUGGCA-GGGCUUCCUGGCCUCCGAGGCCGCGUGUCUUGCUCAUUGCCAGGCAUUCGCUCAUUUUCAAAUCGGAUUUUAUCAAC .........((((--((---....)))-)))..(((.((((.....))))((((((((.((.....)).)))))..))).............))).......... ( -28.40) >DroMoj_CAF1 160070 95 + 1 CUUUUGCCGCCAUCCGC---CGUUCGAUAGGGG-------CGACGCUGCCGCGUGCCUUGCUCAUUGCCAGGCAUUCGCUCAUUUUCAAAUCGGAUUUUAUCAAC ...(((.....(((((.---.(((.((...(((-------((((((....)))(((((.((.....)).))))).))))))....)).)))))))).....))). ( -27.90) >DroPer_CAF1 129664 92 + 1 UUUGUUUCGUCCU--GG-AGCGGUCCUUGAGC----------CUUGAGCCGCGUGUCUUGCUCAUUGCCAGGCAUUCGCUCAUUUUCAAAUCGGAUUUUAUCAAC ...(((((.....--))-)))((((((((((.----------..(((((.(.((((((.((.....)).)))))).))))))..)))))...)))))........ ( -25.70) >consensus UUUUUUUCAUCCU__GC___CGUUCCAUGGGC________C_CCGGAGCCGCGUGUCUUGCUCAUUGCCAGGCAUUCGCUCAUUUUCAAAUCGGAUUUUAUCAAC .............................................((((.(.((((((.((.....)).)))))).)))))........................ (-13.35 = -13.72 + 0.36)

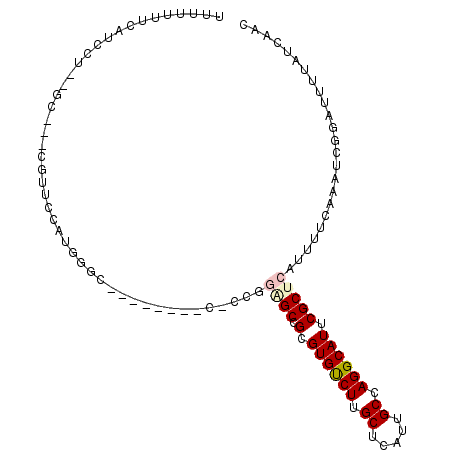
| Location | 2,686,022 – 2,686,130 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.56 |
| Mean single sequence MFE | -27.39 |
| Consensus MFE | -23.56 |
| Energy contribution | -24.06 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938335 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2686022 108 + 22407834 UGCCAGGCAUUCGCUCAUUUUCAAAUCGGAUUUUAUCAACUGAAUGCCAUUCUUGUGCAAAUAUCUUAUCGCGCUGCCG------------CUGCCGCACAGGAUAAUGGCAACUUUUUU (((((((((((((.......((......))..........))))))))..((((((((((.....))...(((....))------------)....))))))))...)))))........ ( -28.13) >DroGri_CAF1 135022 104 + 1 UGCCAGGCAUUCGCUCAUUUUCAAAUCGGAUUUUAUCAACUGAAUGCCAUUCUUGUGCAAAUAUCUUAUCUC----CCG------------UUGGCGCACAGGAUAAUGGCAGCUUUUCC (((((((((((((.......((......))..........))))))))..((((((((..............----...------------.....))))))))...)))))........ ( -26.18) >DroEre_CAF1 120215 114 + 1 UGCCAGGCAUUCGCUCAUUUUCAAAUCGGAUUUUAUCAACUGAAUGCCAUUCUUGUGCAAAUAUCUUAUCUCGCUGCCGCUG------UCGCUGCCGCACAGGAUAAUGGCAACUUUUUU (((((((((((((.......((......))..........))))))))..((((((((((.....))........((.((..------..)).)).))))))))...)))))........ ( -28.73) >DroWil_CAF1 196136 96 + 1 UGCCAGGCAUUCGCUCAUUUUCAAAUCGGAUUUUAUCAACUGAAUGCCAUUCUUGUGCAAAUAUCUUAUCUCGCU------------------------CAGGAUAAUGGCAACUUUUUU (((((((((((((.......((......))..........))))))))..(((((.((..............)).------------------------)))))...)))))........ ( -22.67) >DroYak_CAF1 110059 120 + 1 UGCCAGGCAUUCGCUCAUUUUCAAAUCGGAUUUUAUCAACUGAAUGCCAUUCUUGUGCAAAUAUCUUAUCUCGCUGCCGCUGCCACUGCCGCUGCCGCACAGGAUAAUGGCAACUUUUUU (((((((((((((.......((......))..........))))))))..((((((((((.....)).....((.((.((.......)).)).)).))))))))...)))))........ ( -32.33) >DroAna_CAF1 186667 102 + 1 UGCCAGGCAUUCGCUCAUUUUCAAAUCGGAUUUUAUCAACUGAAUGCCAUUCUUGUGCAAAUAUCUUAUCUCGCU------------------GCCGCACAGGAUAAUGGCAACGUUUCC (((((((((((((.......((......))..........))))))))..((((((((.................------------------...))))))))...)))))........ ( -26.28) >consensus UGCCAGGCAUUCGCUCAUUUUCAAAUCGGAUUUUAUCAACUGAAUGCCAUUCUUGUGCAAAUAUCUUAUCUCGCU_CCG____________CUGCCGCACAGGAUAAUGGCAACUUUUUU (((((((((((((.......((......))..........))))))))..((((((((......................................))))))))...)))))........ (-23.56 = -24.06 + 0.50)

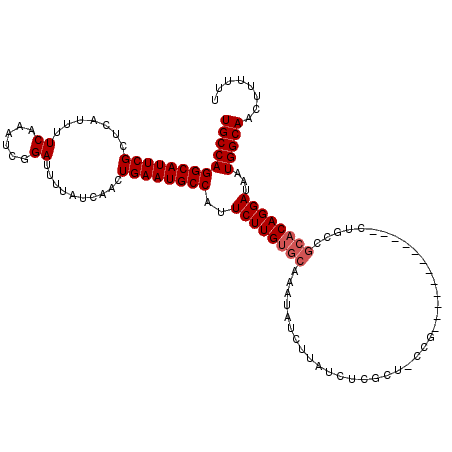
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:49:07 2006