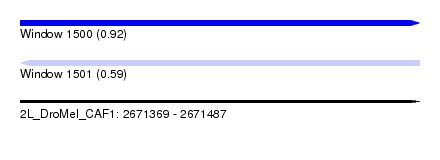
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,671,369 – 2,671,487 |
| Length | 118 |
| Max. P | 0.915643 |

| Location | 2,671,369 – 2,671,487 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 80.73 |
| Mean single sequence MFE | -44.75 |
| Consensus MFE | -33.65 |
| Energy contribution | -37.18 |
| Covariance contribution | 3.53 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915643 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
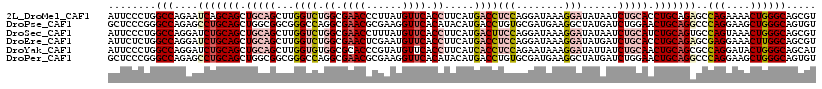
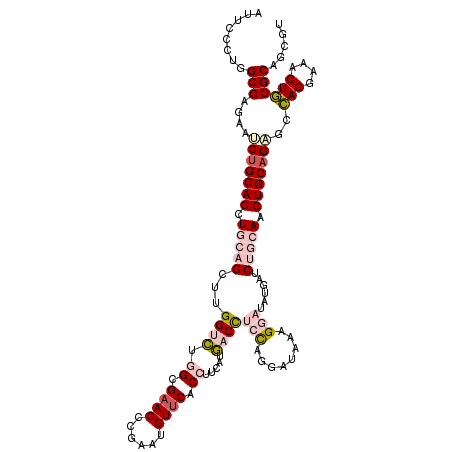
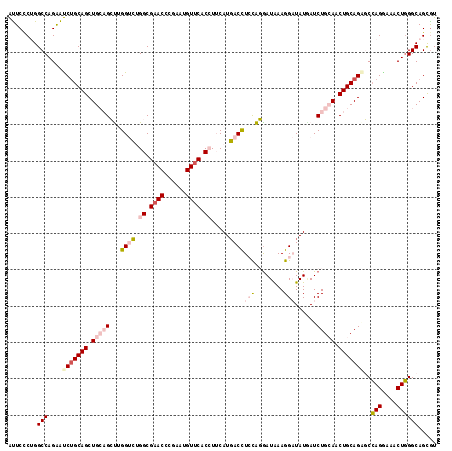
>2L_DroMel_CAF1 2671369 118 + 22407834 ACGCUGCCCAGUUUUCUGGCUCUGCAGGUGCAGAUUAUAUCCUUUAUCCUGGAGGUCAUGAAGGUGAACAUAAGGGUUCGCCAGACCAAGCUGCAGCUGCUGAUUCUGGCCAGGGAAU ..........(((..(((((((.((((.(((((......(((........)))((((.....(((((((......))))))).))))...))))).)))).)).....)))))..))) ( -46.20) >DroPse_CAF1 116226 118 + 1 ACACUGCCCAGCUUCCUGGGCCUGCAGUUCCAGAUCAUAGCCUUCAUCGCACAGGUCAUGUAUGUGAACCUUCGCGUUCGCCUGGCCCGCCGCCAGCUGCAGGCUCUGGCCCGGGAGC ...((.(((.(((....((((((((((............((......(((..(((((((....))).))))..)))...))(((((.....))))))))))))))).)))..))))). ( -44.00) >DroSec_CAF1 91208 118 + 1 ACGCUGCCCAGUUUACUGGCACUGCAGAUGCAGAUUAUAUCCUUUAUCCUGGAAGUCAUGAAGGUGAACAUAAAGGUUCGCCAGACCAAGCUGCAGCUGCAGAUCCUGGCCAGGGAAU ....((((.((....))))))((((((.(((((......(((........))).........(((((((......)))))))........))))).)))))).((((.....)))).. ( -41.20) >DroEre_CAF1 105219 118 + 1 ACGCUGCCAAGUUUCCUCGCUCUGCAGGUGCAGAUCAUAUCCUUUAUCCUGGAGGUCAUGAAGGUGAACAUUCGAGUUCGCCAGACCAAGCUGCAGCUGCAGAUCCUGGCCAGAGAAU .(.((((((.((......))(((((((.(((((.((((..(((((.....)))))..)))).(((((((......)))))))........))))).)))))))...))).))).)... ( -45.00) >DroYak_CAF1 94992 118 + 1 AUGCUGCCCAGUAUCCUGGCGCUGCAGUUGCAGAUAAUAUCCUUUAUUCUGGAGGUGAUGAAGGUGAACAUACGGGUGCGCCACACCAAGCUGCAGCUGCAGAUCCUGGCCAGGGAAU .((((....))))(((((((.((((((((((((......(((........)))((((.....((((.((......)).)))).))))...))))))))))))......)))))))... ( -48.10) >DroPer_CAF1 112966 118 + 1 ACACUGCCCAGCUUCCUGGGCCUGCAGUUCCAGAUCAUAGCCUUCAUCGCACAGGUCAUGUAUGUGAACCUUCGCGUUCGCCUGGCCCGCCGCCAGCUGCAGGCUCUGGCCCGGGAGC ...((.(((.(((....((((((((((............((......(((..(((((((....))).))))..)))...))(((((.....))))))))))))))).)))..))))). ( -44.00) >consensus ACGCUGCCCAGUUUCCUGGCCCUGCAGUUGCAGAUCAUAUCCUUUAUCCUGGAGGUCAUGAAGGUGAACAUACGGGUUCGCCAGACCAAGCUGCAGCUGCAGAUCCUGGCCAGGGAAU ..........(((((((((((((((((.(((((.((((..(((((.....)))))..)))).(((((((......)))))))........))))).))))))).....)))))))))) (-33.65 = -37.18 + 3.53)

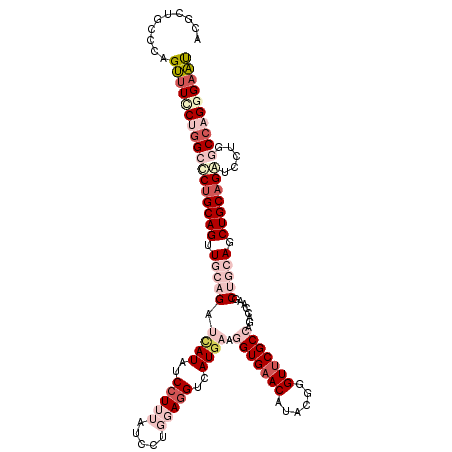
| Location | 2,671,369 – 2,671,487 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 80.73 |
| Mean single sequence MFE | -42.87 |
| Consensus MFE | -27.67 |
| Energy contribution | -29.53 |
| Covariance contribution | 1.87 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.588240 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2671369 118 - 22407834 AUUCCCUGGCCAGAAUCAGCAGCUGCAGCUUGGUCUGGCGAACCCUUAUGUUCACCUUCAUGACCUCCAGGAUAAAGGAUAUAAUCUGCACCUGCAGAGCCAGAAAACUGGGCAGCGU .....(((.((((..(((..(((....))))))((((((((((......)))).((((.((..((....)))).))))......(((((....)))))))))))...)))).)))... ( -37.10) >DroPse_CAF1 116226 118 - 1 GCUCCCGGGCCAGAGCCUGCAGCUGGCGGCGGGCCAGGCGAACGCGAAGGUUCACAUACAUGACCUGUGCGAUGAAGGCUAUGAUCUGGAACUGCAGGCCCAGGAAGCUGGGCAGUGU ((..((((..((.(((((.((.(((((.....))))).....((((.((((.((......)))))).)))).)).))))).))..))))....))..((((((....))))))..... ( -51.20) >DroSec_CAF1 91208 118 - 1 AUUCCCUGGCCAGGAUCUGCAGCUGCAGCUUGGUCUGGCGAACCUUUAUGUUCACCUUCAUGACUUCCAGGAUAAAGGAUAUAAUCUGCAUCUGCAGUGCCAGUAAACUGGGCAGCGU .....(((.((.((..((((((.(((((.((((((.((.((((......)))).)).....))).(((........)))..))).))))).))))))..))((....)))).)))... ( -37.50) >DroEre_CAF1 105219 118 - 1 AUUCUCUGGCCAGGAUCUGCAGCUGCAGCUUGGUCUGGCGAACUCGAAUGUUCACCUUCAUGACCUCCAGGAUAAAGGAUAUGAUCUGCACCUGCAGAGCGAGGAAACUUGGCAGCGU ........(((....(((((((.(((((.((((((.((.((((......)))).)).....))))(((........)))...)).))))).)))))))..(((....))))))..... ( -42.70) >DroYak_CAF1 94992 118 - 1 AUUCCCUGGCCAGGAUCUGCAGCUGCAGCUUGGUGUGGCGCACCCGUAUGUUCACCUUCAUCACCUCCAGAAUAAAGGAUAUUAUCUGCAACUGCAGCGCCAGGAUACUGGGCAGCAU .....(((.((((.((((...(((((((...((((.((.(.((......)).).)).....))))(((........)))............)))))))....)))).)))).)))... ( -37.50) >DroPer_CAF1 112966 118 - 1 GCUCCCGGGCCAGAGCCUGCAGCUGGCGGCGGGCCAGGCGAACGCGAAGGUUCACAUACAUGACCUGUGCGAUGAAGGCUAUGAUCUGGAACUGCAGGCCCAGGAAGCUGGGCAGUGU ((..((((..((.(((((.((.(((((.....))))).....((((.((((.((......)))))).)))).)).))))).))..))))....))..((((((....))))))..... ( -51.20) >consensus AUUCCCUGGCCAGAAUCUGCAGCUGCAGCUUGGUCUGGCGAACCCGAAUGUUCACCUUCAUGACCUCCAGGAUAAAGGAUAUGAUCUGCAACUGCAGAGCCAGGAAACUGGGCAGCGU ........(((....(((((((.(((((...((((.((.((((......)))).)).....))))(((........)))......))))).)))))))..(((....))))))..... (-27.67 = -29.53 + 1.87)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:49:03 2006