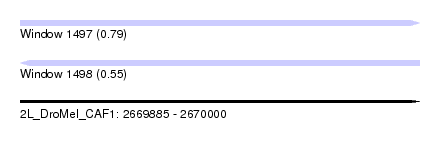
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,669,885 – 2,670,000 |
| Length | 115 |
| Max. P | 0.789438 |

| Location | 2,669,885 – 2,670,000 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.76 |
| Mean single sequence MFE | -32.01 |
| Consensus MFE | -10.87 |
| Energy contribution | -14.27 |
| Covariance contribution | 3.40 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.34 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.789438 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
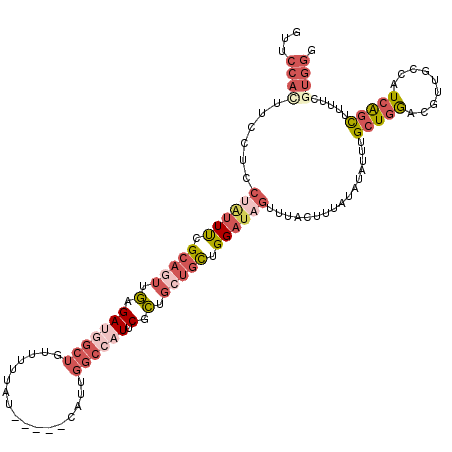
>2L_DroMel_CAF1 2669885 115 + 22407834 GUUCCACUUCCUCCUAUUUCGCAGUUGGGAUGGCUGUUUUUAU-----CAUUGGCCAUUCGCUGCUGUUGGAUAGUUUACUUUAUAUAUUUGCUGGACGUUGCCAUCAGCUUUUCGUGGG ...((((......(((((..(((((.((((((((((.......-----...))))))))).).)))))..)))))................(((((.........))))).....)))). ( -32.00) >DroSim_CAF1 90879 115 + 1 GUUCCACUUCCUCCUAUUUCGCAGUUGGGAUGGCUGUUUUUAU-----CAUUGGCCAUUCGCUGCUGUUGGAUAGUUUACAUUAUAUAUUUGCUGGACGUUGCCGUCAGCUUUUCGUGGG ...((((......(((((..(((((.((((((((((.......-----...))))))))).).)))))..)))))................((((.(((....))))))).....)))). ( -35.70) >DroEre_CAF1 103734 115 + 1 GUGCCACUUCCUCCAAUUUCGCAAUUGAGAUGGCUGCUUUUAU-----CAUUGGCGCUUCGCUGCUGCUGGAUAGCUUACUUUAUAUAUUUGCUGGAGGUUGCCAUCAACUUUUCAUGGG ...(((.......(((((....))))).((((((.(((((...-----..(..((((......)).))..).((((...............))))))))).)))))).........))). ( -26.76) >DroWil_CAF1 170321 115 + 1 GUUCCUUUACCGUCU--UCGGCA---AAGAUAUAUUCUUUUAUAGCAUUCAUUGCAUUGCGUGUCAUCUGGAUUCUAUAUUUUAGCUAUAUGCUAAGCCGAGGCAUCGGUUUUCUGUAUA ........((((.((--((((((---((((.....)))))(((((.(((((.(((((...))).))..))))).)))))..(((((.....))))))))))))...)))).......... ( -29.90) >DroYak_CAF1 93505 115 + 1 GUGCCACUUCCUCCUGUUUCGCAGUGGAGAUGGCUGUUCCUAU-----CAUUGGCCAUUCGCUGCUGCUGGAUAGCUUACAUUAUGUAUUUGCUGGAAGUUGCCAUCAGCUUUUCAUGGG (.((.(((((((((.((...((((((..((((((((.......-----...)))))))))))))).)).))).(((.(((.....)))...))))))))).)))................ ( -35.70) >consensus GUUCCACUUCCUCCUAUUUCGCAGUUGAGAUGGCUGUUUUUAU_____CAUUGGCCAUUCGCUGCUGCUGGAUAGUUUACUUUAUAUAUUUGCUGGACGUUGCCAUCAGCUUUUCGUGGG ...((((......((((((.(((((.(.(((((((.................)))))).).).))))).))))))................(((((.........))))).....)))). (-10.87 = -14.27 + 3.40)

| Location | 2,669,885 – 2,670,000 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.76 |
| Mean single sequence MFE | -26.50 |
| Consensus MFE | -8.55 |
| Energy contribution | -11.71 |
| Covariance contribution | 3.16 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.32 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.554534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
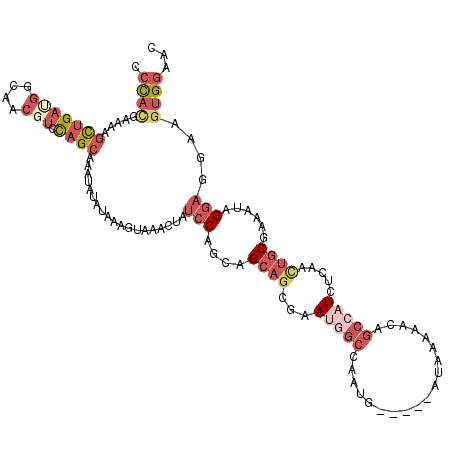
>2L_DroMel_CAF1 2669885 115 - 22407834 CCCACGAAAAGCUGAUGGCAACGUCCAGCAAAUAUAUAAAGUAAACUAUCCAACAGCAGCGAAUGGCCAAUG-----AUAAAAACAGCCAUCCCAACUGCGAAAUAGGAGGAAGUGGAAC .((((.....(((((((....))).))))...................(((....((((.(.(((((.....-----.........))))).)...))))......)))....))))... ( -28.24) >DroSim_CAF1 90879 115 - 1 CCCACGAAAAGCUGACGGCAACGUCCAGCAAAUAUAUAAUGUAAACUAUCCAACAGCAGCGAAUGGCCAAUG-----AUAAAAACAGCCAUCCCAACUGCGAAAUAGGAGGAAGUGGAAC .((((.....(((((((....))).))))...................(((....((((.(.(((((.....-----.........))))).)...))))......)))....))))... ( -30.04) >DroEre_CAF1 103734 115 - 1 CCCAUGAAAAGUUGAUGGCAACCUCCAGCAAAUAUAUAAAGUAAGCUAUCCAGCAGCAGCGAAGCGCCAAUG-----AUAAAAGCAGCCAUCUCAAUUGCGAAAUUGGAGGAAGUGGCAC .((((........((((((........((...(((.....))).(((....))).)).(((...))).....-----.........))))))((((((....)))))).....))))... ( -20.90) >DroWil_CAF1 170321 115 - 1 UAUACAGAAAACCGAUGCCUCGGCUUAGCAUAUAGCUAAAAUAUAGAAUCCAGAUGACACGCAAUGCAAUGAAUGCUAUAAAAGAAUAUAUCUU---UGCCGA--AGACGGUAAAGGAAC ...........((..((((((((((((((.....)))))..(((((.((.((..((.((.....)))).)).)).)))))(((((.....))))---))))))--....))))..))... ( -23.00) >DroYak_CAF1 93505 115 - 1 CCCAUGAAAAGCUGAUGGCAACUUCCAGCAAAUACAUAAUGUAAGCUAUCCAGCAGCAGCGAAUGGCCAAUG-----AUAGGAACAGCCAUCUCCACUGCGAAACAGGAGGAAGUGGCAC .((.......((((..(....)...))))...............(((((...((....))..))))).....-----...))....((((..(((.(((.....)))..)))..)))).. ( -30.30) >consensus CCCACGAAAAGCUGAUGGCAACGUCCAGCAAAUAUAUAAAGUAAACUAUCCAGCAGCAGCGAAUGGCCAAUG_____AUAAAAACAGCCAUCUCAACUGCGAAAUAGGAGGAAGUGGAAC .((((.....(((((((....))).))))...................(((....((((...(((((...................))))).....))))......)))....))))... ( -8.55 = -11.71 + 3.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:49:00 2006