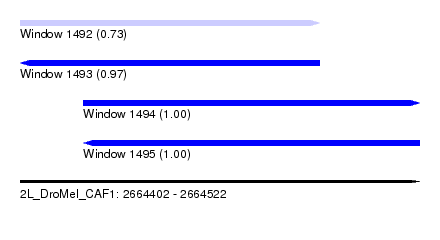
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,664,402 – 2,664,522 |
| Length | 120 |
| Max. P | 0.999846 |

| Location | 2,664,402 – 2,664,492 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 79.57 |
| Mean single sequence MFE | -20.84 |
| Consensus MFE | -10.91 |
| Energy contribution | -12.67 |
| Covariance contribution | 1.76 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.727555 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2664402 90 + 22407834 ACGUGUUCAUAUCCUUGUCAUCCUGCCGGUUGUAUGUGCACACACAUAUGUACAUAU----AUGUACAUAUAUACGGCGCACACACAGUAAGCU ........................((..(((((.(((((.(....(((((((((...----.))))))))).....).))))).)))))..)). ( -21.40) >DroSec_CAF1 83345 78 + 1 ACGUGUUCAUAUCCUUGUCGUCCUGCCGUUUGUGUGUGCACACACAUAUGUA----U----UUGUACA----UACGGC----ACACAGUAAGCU .............((((..((..(((((..((((((....))))))((((((----.----...))))----))))))----).))..)))).. ( -21.20) >DroSim_CAF1 84472 78 + 1 ACGUGUUCAUAUCCUUGUCGUCCUGCCGUUUGUAUGUGCACACACAUAUGUA----U----CUGUACA----UACGGC----ACACAGUAAGCU .............((((..((..((((((.((((((((((........))))----)----..)))))----.)))))----).))..)))).. ( -19.50) >DroEre_CAF1 91735 90 + 1 ACGUGUUCAUAUCCUUGUCGUCCUGCCGUCUGUACGUGUGCACACAUAUACACAUAUACAAUUGUAUAUAUGUACGGC----ACACAGUAAGCU .............((((..((..((((((.((((....)))).(((((((.(((........))).))))))))))))----).))..)))).. ( -20.80) >DroYak_CAF1 87142 74 + 1 ACGUGUUCAUAUCCUUGUCGUCCUGCCGUCUGUAUGUGUGCACAUACA----------------UAUAUGUGCACGGC----ACACAGUAAGCU (((....((......)).)))...((...((((.((((((((((((..----------------..))))))))).))----).))))...)). ( -21.30) >consensus ACGUGUUCAUAUCCUUGUCGUCCUGCCGUUUGUAUGUGCACACACAUAUGUA____U____UUGUACAU_U_UACGGC____ACACAGUAAGCU ..((((.((......)).......(((((.((((((((((......................)))))))))).)))))....))))........ (-10.91 = -12.67 + 1.76)

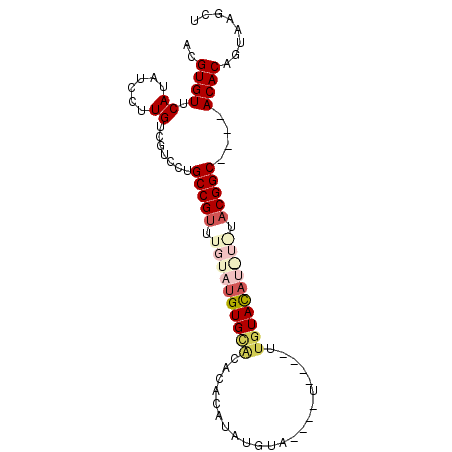
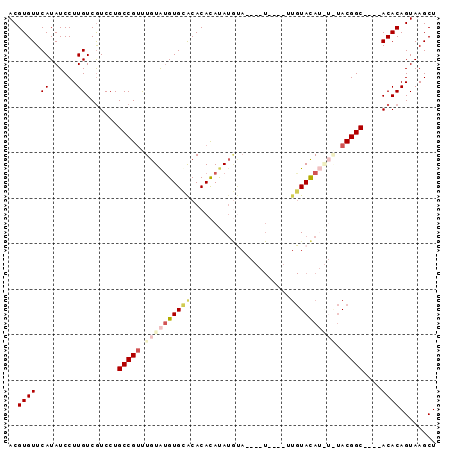
| Location | 2,664,402 – 2,664,492 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 79.57 |
| Mean single sequence MFE | -25.06 |
| Consensus MFE | -13.20 |
| Energy contribution | -14.36 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.974141 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2664402 90 - 22407834 AGCUUACUGUGUGUGCGCCGUAUAUAUGUACAU----AUAUGUACAUAUGUGUGUGCACAUACAACCGGCAGGAUGACAAGGAUAUGAACACGU .(((...(((((((((((.(((((((((((((.----...))))))))))))))))))))))))...)))........................ ( -32.20) >DroSec_CAF1 83345 78 - 1 AGCUUACUGUGU----GCCGUA----UGUACAA----A----UACAUAUGUGUGUGCACACACAAACGGCAGGACGACAAGGAUAUGAACACGU ..(((..(((.(----((((((----((((...----.----))))).((((((....)))))).)))))).).))..)))............. ( -20.90) >DroSim_CAF1 84472 78 - 1 AGCUUACUGUGU----GCCGUA----UGUACAG----A----UACAUAUGUGUGUGCACAUACAAACGGCAGGACGACAAGGAUAUGAACACGU ........((((----.(((((----(((.((.----(----(((....)))).)).))))))...((......))....))......)))).. ( -19.70) >DroEre_CAF1 91735 90 - 1 AGCUUACUGUGU----GCCGUACAUAUAUACAAUUGUAUAUGUGUAUAUGUGUGCACACGUACAGACGGCAGGACGACAAGGAUAUGAACACGU ..(((..(((.(----(((((((((((((((....)))))))))).((((((....))))))...)))))).).))..)))............. ( -27.10) >DroYak_CAF1 87142 74 - 1 AGCUUACUGUGU----GCCGUGCACAUAUA----------------UGUAUGUGCACACAUACAGACGGCAGGACGACAAGGAUAUGAACACGU .(((..((((((----(..(((((((((..----------------..))))))))).)))))))..)))........................ ( -25.40) >consensus AGCUUACUGUGU____GCCGUA_A_AUGUACAA____A____UACAUAUGUGUGUGCACAUACAAACGGCAGGACGACAAGGAUAUGAACACGU ........((((....(((((...((((((............))))))((((((....)))))).))))).(.....)..........)))).. (-13.20 = -14.36 + 1.16)

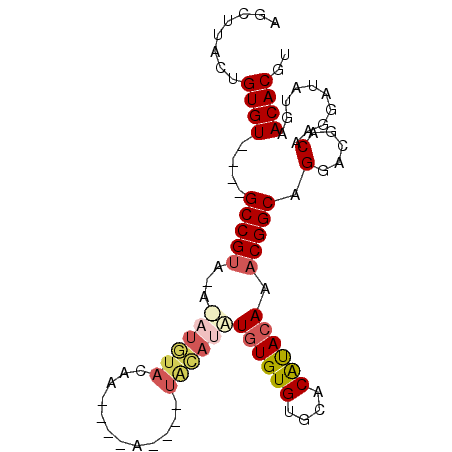
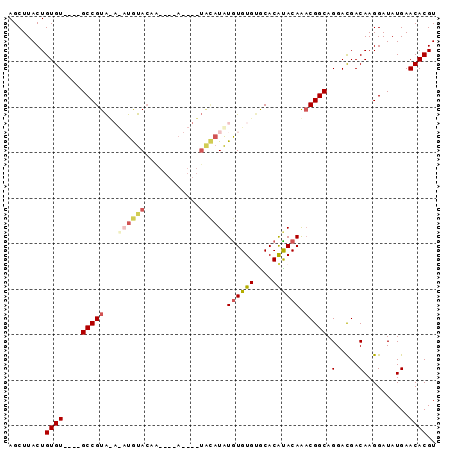
| Location | 2,664,421 – 2,664,522 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 77.39 |
| Mean single sequence MFE | -34.09 |
| Consensus MFE | -22.08 |
| Energy contribution | -22.96 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.18 |
| Structure conservation index | 0.65 |
| SVM decision value | 3.52 |
| SVM RNA-class probability | 0.999340 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2664421 101 + 22407834 AUCCUGCCGGUUGUAUGUGCACACACAUAUGUACAUAU----AUGUACAUAUAUACGGCGCACACACAGUAAGCUCACGCG------CGUGUGUGUGUGCACUGCACUUUG ........(((.(((.(((((((((((((((((((...----.)))))))))(((((.(((.................)))------)))))))))))))))))))))... ( -37.83) >DroSec_CAF1 83364 89 + 1 GUCCUGCCGUUUGUGUGUGCACACACAUAUGUA----U----UUGUACA----UACGGC----ACACAGUAAGCUCACGAG------CGUGUGUGUGUGCACUGCACUUUG ............((((((((((((((((((((.----(----.((..(.----(((...----.....))).)..)).).)------))))))))))))))).)))).... ( -33.50) >DroSim_CAF1 84491 89 + 1 GUCCUGCCGUUUGUAUGUGCACACACAUAUGUA----U----CUGUACA----UACGGC----ACACAGUAAGCUCACGCG------CGUGUGUGUGUGCACUGCACUUUG ...........((((.((((((((((((((((.----.----((((...----.)))).----.........((....)))------)))))))))))))))))))..... ( -32.60) >DroEre_CAF1 91754 97 + 1 GUCCUGCCGUCUGUACGUGUGCACACAUAUACACAUAUACAAUUGUAUAUAUGUACGGC----ACACAGUAAGCUCACGCGU----------GUGUGUGCACUGCACUUUG ....(((.((...(((((((((..(((((((.(((........))).)))))))...))----)))).))).((.((((...----------.)))).)))).)))..... ( -29.80) >DroYak_CAF1 87161 91 + 1 GUCCUGCCGUCUGUAUGUGUGCACAUACA----------------UAUAUGUGCACGGC----ACACAGUAAGCUCACGCGUGUGCGCGUGUGUGUGUGCACUGCACUUUG ....(((.((.......((((((((((..----------------..))))))))))((----((((.....((.((((((....)))))).)))))))))).)))..... ( -36.70) >consensus GUCCUGCCGUUUGUAUGUGCACACACAUAUGUA____U____UUGUACAU_U_UACGGC____ACACAGUAAGCUCACGCG______CGUGUGUGUGUGCACUGCACUUUG ...........((((.(((((((((((((((......................(((............))).((....)).......)))))))))))))))))))..... (-22.08 = -22.96 + 0.88)

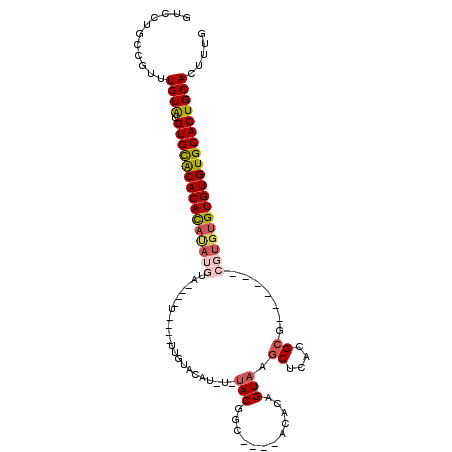
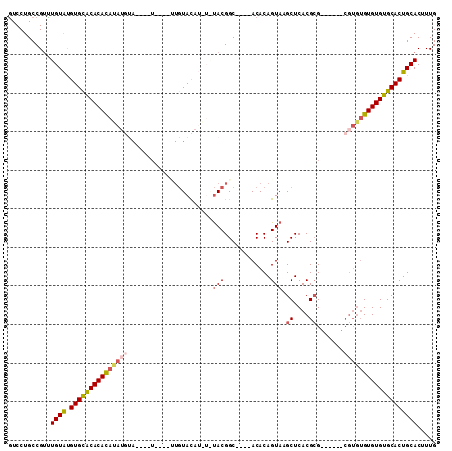
| Location | 2,664,421 – 2,664,522 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 77.39 |
| Mean single sequence MFE | -37.36 |
| Consensus MFE | -19.29 |
| Energy contribution | -20.45 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.19 |
| Mean z-score | -4.40 |
| Structure conservation index | 0.52 |
| SVM decision value | 4.24 |
| SVM RNA-class probability | 0.999846 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2664421 101 - 22407834 CAAAGUGCAGUGCACACACACACG------CGCGUGAGCUUACUGUGUGUGCGCCGUAUAUAUGUACAU----AUAUGUACAUAUGUGUGUGCACAUACAACCGGCAGGAU ....((((((((((((((((....------.((....))....)))))))))))((((((((((((((.----...)))))))))))))))))))......((....)).. ( -44.30) >DroSec_CAF1 83364 89 - 1 CAAAGUGCAGUGCACACACACACG------CUCGUGAGCUUACUGUGU----GCCGUA----UGUACAA----A----UACAUAUGUGUGUGCACACACAAACGGCAGGAC ....(((..((((((((((.((((------(..(((....))).))))----)..(((----((((...----.----))))))))))))))))).)))............ ( -31.50) >DroSim_CAF1 84491 89 - 1 CAAAGUGCAGUGCACACACACACG------CGCGUGAGCUUACUGUGU----GCCGUA----UGUACAG----A----UACAUAUGUGUGUGCACAUACAAACGGCAGGAC .....(((.((((((((((.((((------(((....)).....))))----)..(((----((((...----.----)))))))))))))))))...(....)))).... ( -32.90) >DroEre_CAF1 91754 97 - 1 CAAAGUGCAGUGCACACAC----------ACGCGUGAGCUUACUGUGU----GCCGUACAUAUAUACAAUUGUAUAUGUGUAUAUGUGUGCACACGUACAGACGGCAGGAC .....(((.((((((((((----------(((((((....))).))))----)..((((((((((((....)))))))))))).))))))))).(((....)))))).... ( -39.20) >DroYak_CAF1 87161 91 - 1 CAAAGUGCAGUGCACACACACACGCGCACACGCGUGAGCUUACUGUGU----GCCGUGCACAUAUA----------------UGUAUGUGCACACAUACAGACGGCAGGAC ....((((...)))).....((((((....)))))).(((..((((((----(..(((((((((..----------------..))))))))).)))))))..)))..... ( -38.90) >consensus CAAAGUGCAGUGCACACACACACG______CGCGUGAGCUUACUGUGU____GCCGUA_A_AUGUACAA____A____UACAUAUGUGUGUGCACAUACAAACGGCAGGAC ....(..((((((.(((................))).))..))))..)....(((((...((((((............))))))((((((....)))))).)))))..... (-19.29 = -20.45 + 1.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:48:57 2006