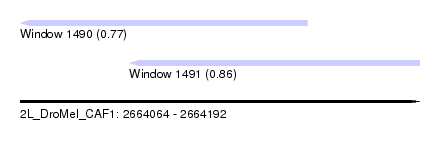
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,664,064 – 2,664,192 |
| Length | 128 |
| Max. P | 0.855980 |

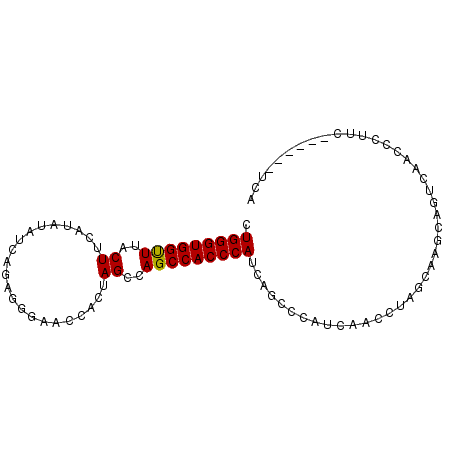
| Location | 2,664,064 – 2,664,156 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 84.25 |
| Mean single sequence MFE | -19.73 |
| Consensus MFE | -13.84 |
| Energy contribution | -13.65 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.771432 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2664064 92 - 22407834 CUGGGUGGCUUACUUCAUAUAUCAGAGGGAACCACUAGCCAGCCACCCAUCAGCCCAUCAACCUAGCAUGCAGUCAACCCUUCAAUCCAUCA .(((((((((..((((........))))............)))))))))........................................... ( -17.54) >DroSec_CAF1 83004 83 - 1 CUGGGUGGUUUACUCCAUAUAUCAGAGGGAACCACUAGCCAGCCACCCAUCAGCCCAUCAACCUGGCAAGCAGUCAACCCUUC--------- ((((((((((..(((.........)))..))))))).(((((....................)))))...)))..........--------- ( -21.35) >DroSim_CAF1 84128 92 - 1 CUGGGUGGUUUACUUCAUAUAUCAGAGGGAACCUCUAGCCAGCCACCCAUCAGCCCAUCAACCUGGCAAGCAGUCAACCCUUCAACCCAUCA .(((((((((.(((.(.......(((((...))))).(((((....................)))))..).))).)))).....)))))... ( -22.75) >DroYak_CAF1 86813 84 - 1 CUGGGUGGUUUACUUCACAUAUCGUAGGUAGCCACUAGCCAGCCACCCAUCAACCCAGCAACCCAUCAAGCAGUCAACCCG--------UCA .((((((((((((((.((.....)))))))((.....)).)))))))))........((..........))..........--------... ( -17.30) >consensus CUGGGUGGUUUACUUCAUAUAUCAGAGGGAACCACUAGCCAGCCACCCAUCAGCCCAUCAACCUAGCAAGCAGUCAACCCUUC______UCA .(((((((((..((......................))..)))))))))........................................... (-13.84 = -13.65 + -0.19)

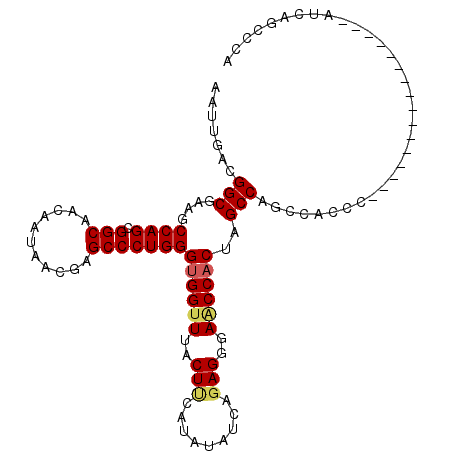
| Location | 2,664,099 – 2,664,192 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 88.13 |
| Mean single sequence MFE | -27.63 |
| Consensus MFE | -23.96 |
| Energy contribution | -24.28 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.855980 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2664099 93 - 22407834 AAUUGACGGCGAAGCCAGCGGCAACAAUAACGAGCCCUGGGUGGCUUACUUCAUAUAUCAGAGGGAACCACUAGCCAGCCACCC----------------AUCAGCCCA .......(((......((.(((...........)))))((((((((..((((........))))............))))))))----------------....))).. ( -25.74) >DroSec_CAF1 83030 93 - 1 AAUUGACGGCGAAGCCAGCGGCAACAAUAACGAGCCCUGGGUGGUUUACUCCAUAUAUCAGAGGGAACCACUAGCCAGCCACCC----------------AUCAGCCCA .......(((....((((.(((...........)))))))((((((..(((.........)))..))))))..)))........----------------......... ( -28.00) >DroSim_CAF1 84163 93 - 1 AAUUGACGGCGAAGCCAGCGGCAACAAUAACGAGCCCUGGGUGGUUUACUUCAUAUAUCAGAGGGAACCUCUAGCCAGCCACCC----------------AUCAGCCCA .......(((......((.(((...........)))))((((((((..((.........(((((...)))))))..))))))))----------------....))).. ( -26.40) >DroEre_CAF1 91416 109 - 1 AAUUGACGGCGAAGCCAGCGGCAACAAUAACGAGCCCUGGGUGGUUUACUCCAUAUAUCAGAGGUAACCACUAGCCAGUCACCCCUCAACCCGUCAACCCAUCAACCCG ..(((((((.(...((((.(((...........)))))))((((((..(((.........)))..))))))..................))))))))............ ( -30.10) >DroYak_CAF1 86840 93 - 1 AAUUGACGGCGAAGCCAGGGGCAACAAUAACGAGCCCUGGGUGGUUUACUUCACAUAUCGUAGGUAGCCACUAGCCAGCCACCC----------------AUCAACCCA ..((((.(((...))).(((((...........)))))(((((((((((((.((.....)))))))((.....)).))))))))----------------.)))).... ( -27.90) >consensus AAUUGACGGCGAAGCCAGCGGCAACAAUAACGAGCCCUGGGUGGUUUACUUCAUAUAUCAGAGGGAACCACUAGCCAGCCACCC________________AUCAGCCCA .......(((....((((.(((...........)))))))((((((..(((.........)))..))))))..)))................................. (-23.96 = -24.28 + 0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:48:54 2006