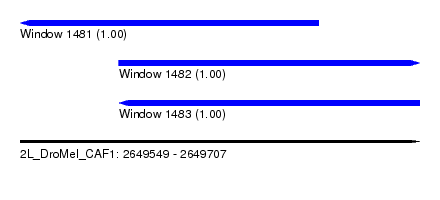
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,649,549 – 2,649,707 |
| Length | 158 |
| Max. P | 0.999987 |

| Location | 2,649,549 – 2,649,667 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 90.76 |
| Mean single sequence MFE | -33.66 |
| Consensus MFE | -30.86 |
| Energy contribution | -30.06 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.12 |
| Structure conservation index | 0.92 |
| SVM decision value | 4.88 |
| SVM RNA-class probability | 0.999959 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2649549 118 - 22407834 UGAAAGUGCUAGUGGUGAAAACCAAUUCAAGAAUGUCAGGUUUUCCCCCUCGCACUUUACCCACAAUAGUUGUUCCUCCGCCCAAAGCAGCUCCGAAAGCAAUUCUUCAAGAAUUCAA ..(((((((.((.((.(((((((....((....))...))))))).)))).))))))).........(((((((...........)))))))......(.(((((.....)))))).. ( -26.60) >DroSec_CAF1 70416 118 - 1 UGAAAGUGCUAGUGGGGAAAACCAAUUCAAGAAUGUCAGGUUUUCCUCCUCGCACUUUACCCACAAUAGUUGUUCCUCCACUCAAAGCAGCUCCGAAAGCAAUUCUUCGAGAAUUCAA ..(((((((.((.((((((((((....((....))...)))))))))))).)))))))..........((((((...........))))))((((((........)))).))...... ( -31.20) >DroSim_CAF1 71459 118 - 1 UGAAAGUGCUAGUGGGGAAAACCAAUUCAAGAAUGUCAGGUUUUCCUCCUCGCACUUUACCCACAAUAGUUGUUCCUCCACUCAAAGCAGCUCCGAAAGCAAUUCUUCAAGAAUUCAA ..(((((((.((.((((((((((....((....))...)))))))))))).))))))).........(((((((...........)))))))......(.(((((.....)))))).. ( -30.70) >DroEre_CAF1 78812 118 - 1 UGAAAGUGCUAGUGGGGAAAACCAAUUCAGAAAUGUCAGGUUUUCCCCCUCGCACUUUGGCCACAAAAGUUGUUCCUCUACCCAAAACAGCUCCGAAGGCAGUUCUUCAAGAGUUCCA ..(((((((.((.((((((((((....((....))...)))))))))))).))))))).(((.....(((((((...........))))))).....))).................. ( -40.00) >DroYak_CAF1 73861 118 - 1 UGAAAGUGCUAGUGGGGAAAAUCAAUUCAGAAAUGUCAGGUUUUCCCCCUCGCACUUUGACCACAAAAGUGGUUCCUCUACCGAAAACAGCUCCGCAAGCAGUUUGUCAAGAAUUCAA ..(((((((.((.((((((((((....((....))...)))))))))))).)))))))((((((....))))))........((....((((.(....).))))..)).......... ( -39.80) >consensus UGAAAGUGCUAGUGGGGAAAACCAAUUCAAGAAUGUCAGGUUUUCCCCCUCGCACUUUACCCACAAUAGUUGUUCCUCCACCCAAAGCAGCUCCGAAAGCAAUUCUUCAAGAAUUCAA ..(((((((.((.((((((((((....((....))...)))))))))))).))))))).........(((((((...........)))))))......(.(((((.....)))))).. (-30.86 = -30.06 + -0.80)

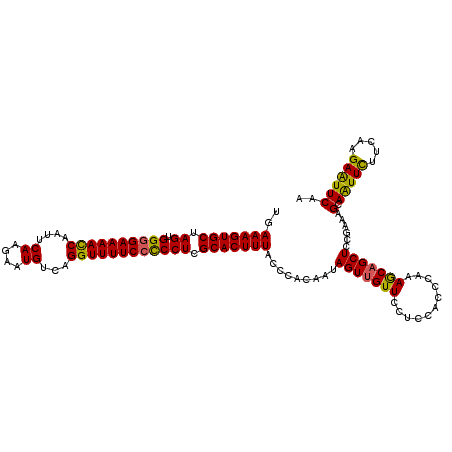
| Location | 2,649,588 – 2,649,707 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 90.67 |
| Mean single sequence MFE | -37.67 |
| Consensus MFE | -29.88 |
| Energy contribution | -31.88 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.16 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.73 |
| SVM RNA-class probability | 0.996693 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2649588 119 + 22407834 GGAGGAACAACUAUUGUGGGUAAAGUGCGAGGGGGAAAACCUGACAUUCUUGAAUUGGUUUUCACCACUAGCACUUUCAAAUAUUGUGCUUCCACUACUUUGUCUCGAUACUGAACGAA (((((.((((.((((....(.(((((((.((((.(((((((...((....))....))))))).)).)).)))))))).)))))))).)))))......((((.(((....))))))). ( -34.00) >DroSec_CAF1 70455 119 + 1 GGAGGAACAACUAUUGUGGGUAAAGUGCGAGGAGGAAAACCUGACAUUCUUGAAUUGGUUUUCCCCACUAGCACUUUCACAAAUUAUGCUUCCACUACUUUGUCUCGGUUCUGAACGAA (((((........((((((...((((((..((.((((((((...((....))....))))))))))....))))))))))))......)))))...........(((........))). ( -34.34) >DroSim_CAF1 71498 119 + 1 GGAGGAACAACUAUUGUGGGUAAAGUGCGAGGAGGAAAACCUGACAUUCUUGAAUUGGUUUUCCCCACUAGCACUUUCACAAAUUGUGCUUCCACUACUUUGUCUCGGUUCUAAACGAA (((((.((((...((((((...((((((..((.((((((((...((....))....))))))))))....)))))))))))).)))).)))))...........(((........))). ( -37.80) >DroEre_CAF1 78851 119 + 1 AGAGGAACAACUUUUGUGGCCAAAGUGCGAGGGGGAAAACCUGACAUUUCUGAAUUGGUUUUCCCCACUAGCACUUUCACAAAUUGUGCUUCCACUACUUUGUCUCAAUGCAGAAAGAU ...((((((...(((((((...((((((.((((((((((((...............)))))))))).)).)))))))))))))...)).)))).....(((((......)))))..... ( -41.16) >DroYak_CAF1 73900 119 + 1 AGAGGAACCACUUUUGUGGUCAAAGUGCGAGGGGGAAAACCUGACAUUUCUGAAUUGAUUUUCCCCACUAGCACUUUCACAAAUUGUGCUUCCACUACUUUGUCUCAAUUCUGAUAGAG ...((((.(((.(((((((...((((((.((((((((((.(...............).)))))))).)).)))))))))))))..))).))))....(((((((........))))))) ( -41.06) >consensus GGAGGAACAACUAUUGUGGGUAAAGUGCGAGGGGGAAAACCUGACAUUCUUGAAUUGGUUUUCCCCACUAGCACUUUCACAAAUUGUGCUUCCACUACUUUGUCUCGAUUCUGAACGAA (((((.((((...((((((...((((((.((((((((((((...((....))....)))))))))).)).)))))))))))).)))).))))).......................... (-29.88 = -31.88 + 2.00)

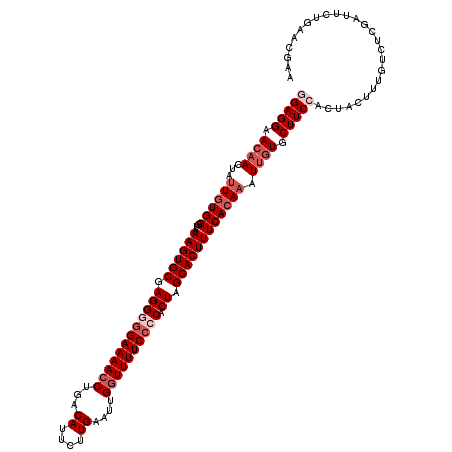
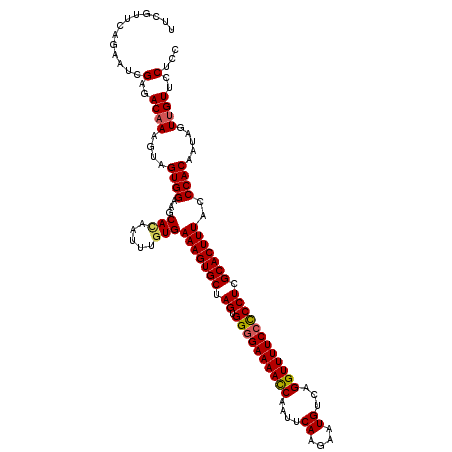
| Location | 2,649,588 – 2,649,707 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 90.67 |
| Mean single sequence MFE | -37.36 |
| Consensus MFE | -35.48 |
| Energy contribution | -35.56 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.28 |
| Structure conservation index | 0.95 |
| SVM decision value | 5.45 |
| SVM RNA-class probability | 0.999987 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2649588 119 - 22407834 UUCGUUCAGUAUCGAGACAAAGUAGUGGAAGCACAAUAUUUGAAAGUGCUAGUGGUGAAAACCAAUUCAAGAAUGUCAGGUUUUCCCCCUCGCACUUUACCCACAAUAGUUGUUCCUCC .............(..((((....((((.....((.....))(((((((.((.((.(((((((....((....))...))))))).)))).)))))))..)))).....))))..)... ( -28.90) >DroSec_CAF1 70455 119 - 1 UUCGUUCAGAACCGAGACAAAGUAGUGGAAGCAUAAUUUGUGAAAGUGCUAGUGGGGAAAACCAAUUCAAGAAUGUCAGGUUUUCCUCCUCGCACUUUACCCACAAUAGUUGUUCCUCC .............(..((((....((((..(((.....))).(((((((.((.((((((((((....((....))...)))))))))))).)))))))..)))).....))))..)... ( -35.20) >DroSim_CAF1 71498 119 - 1 UUCGUUUAGAACCGAGACAAAGUAGUGGAAGCACAAUUUGUGAAAGUGCUAGUGGGGAAAACCAAUUCAAGAAUGUCAGGUUUUCCUCCUCGCACUUUACCCACAAUAGUUGUUCCUCC .............(..((((....((((...(((.....)))(((((((.((.((((((((((....((....))...)))))))))))).)))))))..)))).....))))..)... ( -36.40) >DroEre_CAF1 78851 119 - 1 AUCUUUCUGCAUUGAGACAAAGUAGUGGAAGCACAAUUUGUGAAAGUGCUAGUGGGGAAAACCAAUUCAGAAAUGUCAGGUUUUCCCCCUCGCACUUUGGCCACAAAAGUUGUUCCUCU .............(((((((....(((....)))..(((((((((((((.((.((((((((((....((....))...)))))))))))).)))))))...))))))..))))..))). ( -42.30) >DroYak_CAF1 73900 119 - 1 CUCUAUCAGAAUUGAGACAAAGUAGUGGAAGCACAAUUUGUGAAAGUGCUAGUGGGGAAAAUCAAUUCAGAAAUGUCAGGUUUUCCCCCUCGCACUUUGACCACAAAAGUGGUUCCUCU (((..........))).......((.((((.(((..(((((((((((((.((.((((((((((....((....))...)))))))))))).)))))))...)))))).))).)))).)) ( -44.00) >consensus UUCGUUCAGAAUCGAGACAAAGUAGUGGAAGCACAAUUUGUGAAAGUGCUAGUGGGGAAAACCAAUUCAAGAAUGUCAGGUUUUCCCCCUCGCACUUUACCCACAAUAGUUGUUCCUCC .............(..((((....((((...(((.....)))(((((((.((.((((((((((....((....))...)))))))))))).)))))))..)))).....))))..)... (-35.48 = -35.56 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:48:46 2006