| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,640,778 – 2,640,870 |
| Length | 92 |
| Max. P | 0.826944 |

| Location | 2,640,778 – 2,640,870 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.01 |
| Mean single sequence MFE | -31.00 |
| Consensus MFE | -19.58 |
| Energy contribution | -20.54 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.561753 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2640778 92 + 22407834 CCACAACGCAAACAUAAGCAGGUGCACCGCGGCUGC----------------------------UCCUAUGGAACAGCUCCUUUGGAACACUUCCAGGAGUGCCCACUAAUGUGUUUCUU ....((((((......(((((.(((...))).))))----------------------------)....(((....((((((..(((.....)))))))))..)))....)))))).... ( -27.30) >DroSec_CAF1 61426 92 + 1 CCACAACGCACACAUAAGCAGGUGCACCGCGGCAGC----------------------------UCCUGUGGAACAGCUCAUUUGGAACACUUCCAGGAGUGCCCACUAAUGUGUUUCUU .........((((((.((..((.((.((((((....----------------------------..))))))....((((...((((.....)))).)))))))).)).))))))..... ( -26.50) >DroSim_CAF1 62524 92 + 1 CCACAACGCACACAUAAGCAGGUUCACCGCGGCAGC----------------------------UCCUGUGGAACACCUCCUUUGGAACACUUCCAGGAGUGCCCACUAAUGUGUUUCUU .........((((((..((.((....))))((((.(----------------------------(((((.(((...((......)).....))))))))))))).....))))))..... ( -27.50) >DroEre_CAF1 69809 120 + 1 CCAAGCCGCACACAUAAGCAGGUGCACCACGGCAGCUCCUGUGGAACAGCUCCUGUGGAACAGCUCCUGUGGAACAGCUCCUUUGGAACACUUGCAGGAGUGCCCACUAAUGUGUUUCUU .........((((((.((..((.((.((((((......))))))....(((((((..(....(.(((...(((.....)))...))).)..)..))))))))))).)).))))))..... ( -39.00) >DroYak_CAF1 63885 100 + 1 CCACGGUGCACACAUAAGCAGGUGCACCACGGCAGC--------------------GGAACAGCUCCUUUGGAACAGCUCCUUGGGAACACUUCCAGGAGUGCCCACUAAUGUGUUUCUU .((((((((((.(.......))))))))..((.(((--------------------......)))))..(((....((((((.((((....))))))))))..))).....)))...... ( -34.70) >consensus CCACAACGCACACAUAAGCAGGUGCACCGCGGCAGC____________________________UCCUGUGGAACAGCUCCUUUGGAACACUUCCAGGAGUGCCCACUAAUGUGUUUCUU .........((((((.((..(((...((((((..................................))))))....((((((..(((.....))))))))))))..)).))))))..... (-19.58 = -20.54 + 0.96)
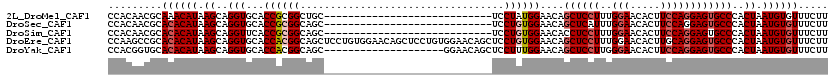
| Location | 2,640,778 – 2,640,870 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.01 |
| Mean single sequence MFE | -37.30 |
| Consensus MFE | -24.98 |
| Energy contribution | -25.34 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.826944 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
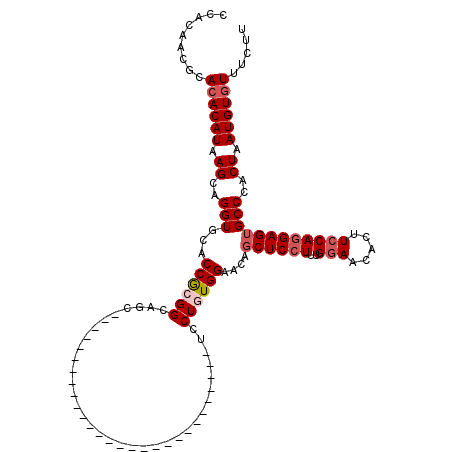
>2L_DroMel_CAF1 2640778 92 - 22407834 AAGAAACACAUUAGUGGGCACUCCUGGAAGUGUUCCAAAGGAGCUGUUCCAUAGGA----------------------------GCAGCCGCGGUGCACCUGCUUAUGUUUGCGUUGUGG ......((((...(..((((.(((((((.....)))..))))((((((((...)))----------------------------))))).((((.....))))...))))..)..)))). ( -32.10) >DroSec_CAF1 61426 92 - 1 AAGAAACACAUUAGUGGGCACUCCUGGAAGUGUUCCAAAUGAGCUGUUCCACAGGA----------------------------GCUGCCGCGGUGCACCUGCUUAUGUGUGCGUUGUGG ......(((....)))(((((((((((((..((((.....))))..))))..))))----------------------------).))))((..((((((.......).)))))..)).. ( -32.60) >DroSim_CAF1 62524 92 - 1 AAGAAACACAUUAGUGGGCACUCCUGGAAGUGUUCCAAAGGAGGUGUUCCACAGGA----------------------------GCUGCCGCGGUGAACCUGCUUAUGUGUGCGUUGUGG .....((((((.(((.((((((((((.......(((...)))((....)).)))))----------------------------).))))..((....)).))).))))))......... ( -31.10) >DroEre_CAF1 69809 120 - 1 AAGAAACACAUUAGUGGGCACUCCUGCAAGUGUUCCAAAGGAGCUGUUCCACAGGAGCUGUUCCACAGGAGCUGUUCCACAGGAGCUGCCGUGGUGCACCUGCUUAUGUGUGCGGCUUGG (((..((((((.(((((((((..(.(((...(((((...))))).(((((...(((((.(((((...))))).)))))...)))))))).)..)))).)).))).))))))....))).. ( -48.60) >DroYak_CAF1 63885 100 - 1 AAGAAACACAUUAGUGGGCACUCCUGGAAGUGUUCCCAAGGAGCUGUUCCAAAGGAGCUGUUCC--------------------GCUGCCGUGGUGCACCUGCUUAUGUGUGCACCGUGG ......(((..((((((((((((((((((..(((((...)))))..))))..))))).)).)))--------------------))))....((((((((.......).)))))))))). ( -42.10) >consensus AAGAAACACAUUAGUGGGCACUCCUGGAAGUGUUCCAAAGGAGCUGUUCCACAGGA____________________________GCUGCCGCGGUGCACCUGCUUAUGUGUGCGUUGUGG .....((((((.((..(((((((((((((..((((.....))))..))))..))))............................((....)).)))..))..)).))))))......... (-24.98 = -25.34 + 0.36)

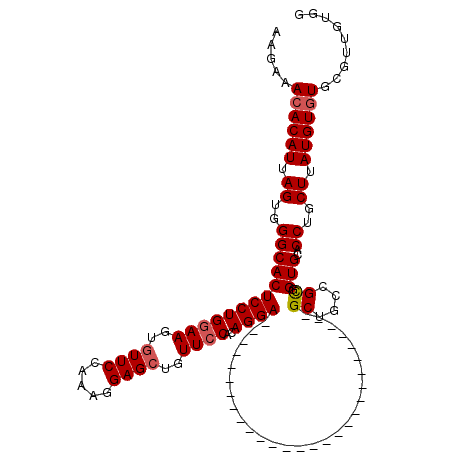
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:48:41 2006