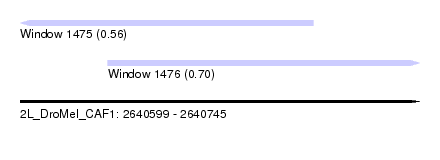
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,640,599 – 2,640,745 |
| Length | 146 |
| Max. P | 0.698835 |

| Location | 2,640,599 – 2,640,706 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.27 |
| Mean single sequence MFE | -35.31 |
| Consensus MFE | -20.79 |
| Energy contribution | -21.27 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.556587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
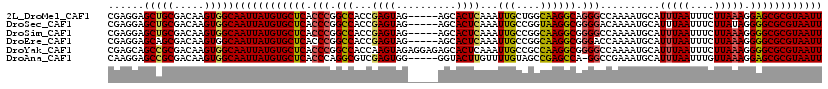
>2L_DroMel_CAF1 2640599 107 - 22407834 CUGCCUUGCCAGCAAUUUGAGUGCU-----CUACUCGGUGGCCGGGUGAGCACAUAAUUGCCACUUGUCGCAGCUCCUCGAGAAACGCU--UUCUUGGCGUU------CUUCAACAAAAA ((((.......((((((...(((((-----(.((((((...))))))))))))..))))))........))))......((..((((((--.....))))))------..))........ ( -36.66) >DroSec_CAF1 61251 103 - 1 CCGCCUUACCGGCAAUUUGAGUGCU-----CUACUCGGUGGCCGGGUGAGCACAUAAUUGCCACUUGUCGCAGCUCCUCGAGAAGCGCU--UCCUUGGCGUU------CUUCAACA---- .((((.....(((((((...(((((-----(.((((((...))))))))))))..)))))))....(.(((..(((...)))..)))).--.....))))..------........---- ( -39.30) >DroSim_CAF1 62345 107 - 1 CCGCCUUGCCGGCAAUUUGAGUGCU-----CUACUCGGUGGCCGGGUGAGCACAUAAUUGCCACUUGUCGCAGCUCCUCGAGAAACGCU--UCCUUGGCGUU------CUUCUACAAUAU ..((......(((((((...(((((-----(.((((((...))))))))))))..))))))).......))........((..((((((--.....))))))------..))........ ( -37.02) >DroEre_CAF1 69631 100 - 1 CCGCCUUGCCGGCAAUUUGAGUGCU-----CUACUCGGUGGCCGGGUGAGCACAUAAUUGCCACUUGUCGCUGCUCCUCGAGAAACGCU--UCCUUGCCGCU------CUCCA------- ..........(((((((...(((((-----(.((((((...))))))))))))..))))))).................((((...((.--.....))...)------)))..------- ( -33.00) >DroYak_CAF1 63700 113 - 1 CCGCCUUGGCGGCAAUUUGAGUGCUCUCCUCUACUUGGUGGCCGGGUGAGCACAUAAUUGCCACUUGUCGCGGCUGCUCGAGAAACGCCGCUCCUUGCCGCUCCUUCUCUCCA------- ((((...((.(((((((...((((((.(((((((...))))..))).))))))..))))))).))....))))......(((((..((.((.....)).))...)))))....------- ( -42.00) >DroAna_CAF1 83653 99 - 1 -UGGCUCGGCUACAAAACAAGUACC-----CCACUCGACGCCUGGGUGAGCACAUAAUUGCCACUUGUCGCGGCUCCUUGAGAAAUGCU--UCUUCGGCGCU------CUCAA------- -.((...(((..(...((((((...-----.((((((.....)))))).(((......))).)))))).)..))))).(((((..((((--.....)))).)------)))).------- ( -23.90) >consensus CCGCCUUGCCGGCAAUUUGAGUGCU_____CUACUCGGUGGCCGGGUGAGCACAUAAUUGCCACUUGUCGCAGCUCCUCGAGAAACGCU__UCCUUGGCGCU______CUCCA_______ ....((((..(((....((((((........))))))(((((.(((((.(((......))))))))))))).)))...))))...((((.......)))).................... (-20.79 = -21.27 + 0.48)

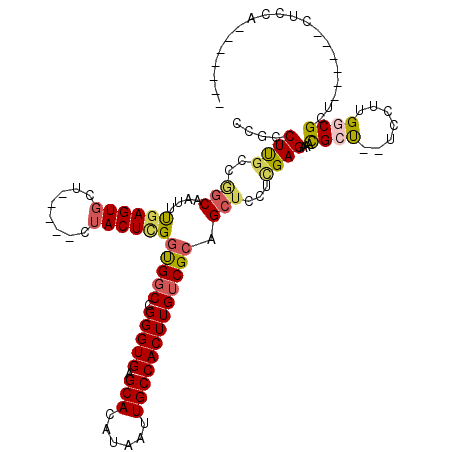
| Location | 2,640,631 – 2,640,745 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 89.11 |
| Mean single sequence MFE | -38.37 |
| Consensus MFE | -32.92 |
| Energy contribution | -32.70 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.698835 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2640631 114 + 22407834 CGAGGAGCUGCGACAAGUGGCAAUUAUGUGCUCACCCGGCCACCGAGUAG-----AGCACUCAAAUUGCUGGCAAGGCAGGGCCAAAAUGCAUUUAAUUUCUUAAAGGAGCGCGUAAUU ......((..(.....)..))((((((((((((....((((...((((..-----...))))...(((((.....)))))))))........(((((....))))).)))))))))))) ( -37.80) >DroSec_CAF1 61279 114 + 1 CGAGGAGCUGCGACAAGUGGCAAUUAUGUGCUCACCCGGCCACCGAGUAG-----AGCACUCAAAUUGCCGGUAAGGCGGGGACAAAAUGCAUUUAAUUUCUUAUAGGGGCGCGUAAUU ......((..(.....)..))((((((((((((.(((.(((((((.((((-----..........))))))))..))).)))...((((.......)))).......)))))))))))) ( -37.20) >DroSim_CAF1 62377 114 + 1 CGAGGAGCUGCGACAAGUGGCAAUUAUGUGCUCACCCGGCCACCGAGUAG-----AGCACUCAAAUUGCCGGCAAGGCGGGGCCAAAAUGCAUUUAAUUUCUUAAAGGGGCGCGUAAUU ......((..(.....)..))((((((((((((....((((.((((((..-----...)))).....(((.....)))))))))........(((((....))))).)))))))))))) ( -41.00) >DroEre_CAF1 69656 114 + 1 CGAGGAGCAGCGACAAGUGGCAAUUAUGUGCUCACCCGGCCACCGAGUAG-----AGCACUCAAAUUGCCGGCAAGGCGGGACCAAAAUGCAUUUAAUUUCUUAAAGGGGCGCGUAAUU ......((.((.....)).))((((((((((((.((((.((...((((..-----...))))...(((....)))))))))...........(((((....))))).)))))))))))) ( -32.00) >DroYak_CAF1 63733 119 + 1 CGAGCAGCCGCGACAAGUGGCAAUUAUGUGCUCACCCGGCCACCAAGUAGAGGAGAGCACUCAAAUUGCCGCCAAGGCGGGGCCAAAAUGCAUUUAAUUUCUUAAAGGGGCGCGUAAUU ......(((((.....)))))((((((((((((....((((.((.....(((.......))).....(((.....)))))))))........(((((....))))).)))))))))))) ( -43.30) >DroAna_CAF1 83678 113 + 1 CAAGGAGCCGCGACAAGUGGCAAUUAUGUGCUCACCCAGGCGUCGAGUGG-----GGUACUUGUUUUGUAGCCGAGCCA-GGCCGAAAUGCAUUUAAUUUGUUAAAGGAGCGCGUAAUU ......(((((.....)))))((((((((((((((((..((.....)).)-----))).(((((((((..(((......-)))))))))(((.......)))..))))))))))))))) ( -38.90) >consensus CGAGGAGCUGCGACAAGUGGCAAUUAUGUGCUCACCCGGCCACCGAGUAG_____AGCACUCAAAUUGCCGGCAAGGCGGGGCCAAAAUGCAUUUAAUUUCUUAAAGGGGCGCGUAAUU ......(((((.....)))))((((((((((((.(((.(((...((((..........))))...(((....)))))).)))..........(((((....))))).)))))))))))) (-32.92 = -32.70 + -0.22)
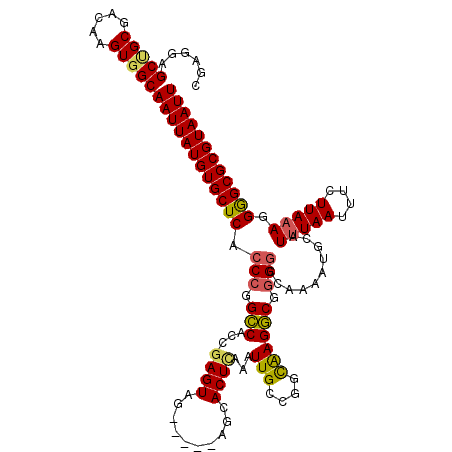
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:48:39 2006