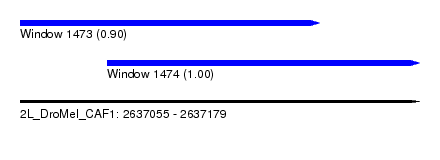
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,637,055 – 2,637,179 |
| Length | 124 |
| Max. P | 0.998712 |

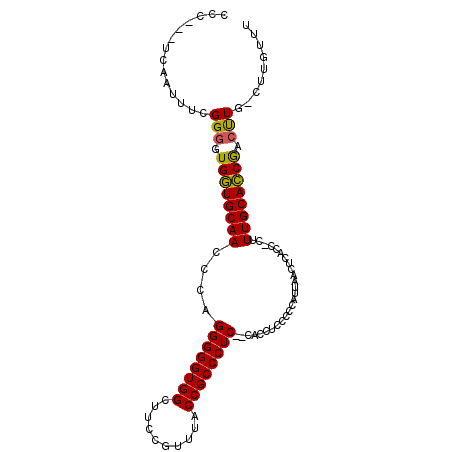
| Location | 2,637,055 – 2,637,148 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 80.89 |
| Mean single sequence MFE | -29.55 |
| Consensus MFE | -19.40 |
| Energy contribution | -19.80 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.902708 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
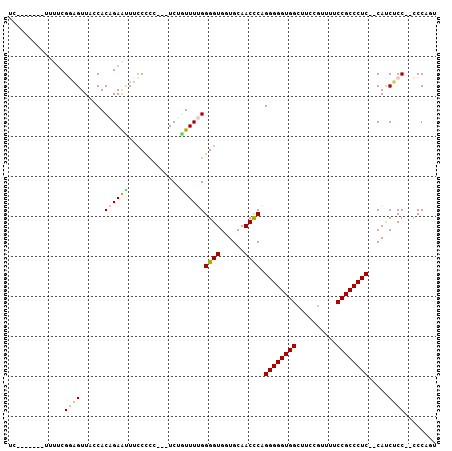
>2L_DroMel_CAF1 2637055 93 + 22407834 UC-------UUUUCGUUGUUACCACAGAACUUGCCAC---UCUGUUUUGGGGUGGUGCAACCCAGGGGGUGGCUUCCUUUUUCCGCCCUC--CAUCACC--CCCAGU ..-------..............(((((.........---)))))...(((((((((.......((((((((..........))))))))--)))))))--)).... ( -29.61) >DroSec_CAF1 57704 95 + 1 UC-------UUUUCGGAGUUAACACAGAAUUUCCUCC---UCUGUUUUGGGGUGCUGCAACCCAGGGGGUGGCUUCCGUUUUCCGCCCUC--CAUCUCCCCCCCUGU ..-------.....((((.(((.(((((.........---))))).)))((((......)))).((((((((..........)))))).)--)..))))........ ( -27.20) >DroSim_CAF1 58783 94 + 1 UC-------UUUUCGGAGUUAACACAGAAUUUCCUCC---UCUGUUUUGGGGUGGUGCAACCCAGGGGGUGGCUUCCGUUUUCCGCCCUC--CAUCGCC-CCCCUGU ..-------.....((((..((.......))..))))---........(((((((((.......((((((((..........))))))))--)))))))-))..... ( -30.81) >DroEre_CAF1 66082 93 + 1 UU-------UUCCCGGAGUUACCACAGAAUUUCCCCC---UCAAUUUCGGGGUGGUGCAACUCAGGGGGUGGCUUCCGCUUGCCGCCCUC--CACCUCC--CCCAUU ..-------.....((((............))))...---........((((.((((.......(((((((((........)))))))))--)))).))--)).... ( -33.31) >DroYak_CAF1 60135 105 + 1 CUUGCCACUUUUUCGUAGUUACCACAGAGUUUCCCCCUCCUCAUUUUGGGGGUGUUGCAACCCAGGGGGUGGCUUCCGUUUACCGCCCUCCCCACCUCC--GCCAUU .((((.(((((...((....))...)))))..(((((..........)))))....))))....((((((((.....((.....)).....))))))))--...... ( -26.80) >consensus UC_______UUUUCGGAGUUACCACAGAAUUUCCCCC___UCUGUUUUGGGGUGGUGCAACCCAGGGGGUGGCUUCCGUUUUCCGCCCUC__CAUCUCC__CCCAGU ..............((((......((((((.............))))))((((......)))).((((((((..........)))))))).....))))........ (-19.40 = -19.80 + 0.40)

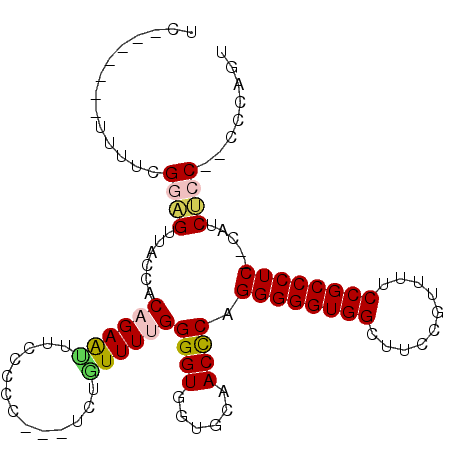
| Location | 2,637,082 – 2,637,179 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 77.56 |
| Mean single sequence MFE | -30.98 |
| Consensus MFE | -21.66 |
| Energy contribution | -21.11 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.70 |
| SVM decision value | 3.20 |
| SVM RNA-class probability | 0.998712 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2637082 97 + 22407834 CAC---UCUGUUUUGGGGUGGUGCAACCCAGGGGGUGGCUUCCUUUUUCCGCCCUC--CAUCACCCCCAGUGAUUCCCC-CCUUGCACCGAACUUGCUUGUUU .((---(((.....)))))((((((....((((((.(((...........)))...--.(((((.....)))))..)))-)))))))))((((......)))) ( -31.50) >DroEre_CAF1 66109 96 + 1 CCC---UCAAUUUCGGGGUGGUGCAACUCAGGGGGUGGCUUCCGCUUGCCGCCCUC--CACCUCCCCCAUUAACCCACU-CUUUGCACCGACUUG-CUUGUUU ...---........((((.((((.......(((((((((........)))))))))--)))).))))............-....(((......))-)...... ( -33.01) >DroYak_CAF1 60169 103 + 1 CCCUCCUCAUUUUGGGGGUGUUGCAACCCAGGGGGUGGCUUCCGUUUACCGCCCUCCCCACCUCCGCCAUUAACUCACCCCUUUGCAACAACUUGUCUCGUUU .(((((.......)))))((((((((....((((((((.....(((....((.............))....)))))))))))))))))))............. ( -28.42) >consensus CCC___UCAAUUUCGGGGUGGUGCAACCCAGGGGGUGGCUUCCGUUUACCGCCCUC__CACCUCCCCCAUUAACUCACC_CUUUGCACCGACUUG_CUUGUUU ..............(((.((((((((....((((((((..........))))))))..........................)))))))).)))......... (-21.66 = -21.11 + -0.55)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:48:37 2006