| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,629,407 – 2,629,539 |
| Length | 132 |
| Max. P | 0.924109 |

| Location | 2,629,407 – 2,629,502 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 86.72 |
| Mean single sequence MFE | -25.10 |
| Consensus MFE | -19.12 |
| Energy contribution | -19.02 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713544 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
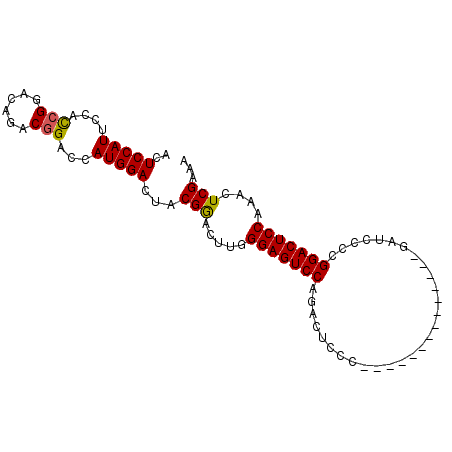
>2L_DroMel_CAF1 2629407 95 + 22407834 ACUCCAUUCCAUCGGACGGACAGACCAUGGACUACGAACUUGGGAGUCCAGACUCCCGAACCCCGAACCCCGAUCCCCGGACUCCAAACUCGAAA .......(((((.((.(.....).)))))))...(((..((((((((....))))))))..........(((.....))).........)))... ( -23.70) >DroSec_CAF1 50119 81 + 1 ACUCCAUUCCACCGGACAGACGGACCAUGGACUACGGACUUGGGAGUCCAGACUCCC--------------GAUCCCCGGACUCCAAACUCGAAA ......(((..(((......)))....((((...(((..((((((((....))))))--------------))...)))...)))).....))). ( -25.80) >DroSim_CAF1 50184 81 + 1 ACUCCAUUCCACCGGACAGACGGACCAUGGACUACGGACUUGGGAGUCCAGACUCCC--------------GAUCCCCGGACUCCAAACUCGAAA ......(((..(((......)))....((((...(((..((((((((....))))))--------------))...)))...)))).....))). ( -25.80) >consensus ACUCCAUUCCACCGGACAGACGGACCAUGGACUACGGACUUGGGAGUCCAGACUCCC______________GAUCCCCGGACUCCAAACUCGAAA ..(((((....(((......)))...)))))...(((.....(((((((.............................)))))))....)))... (-19.12 = -19.02 + -0.11)

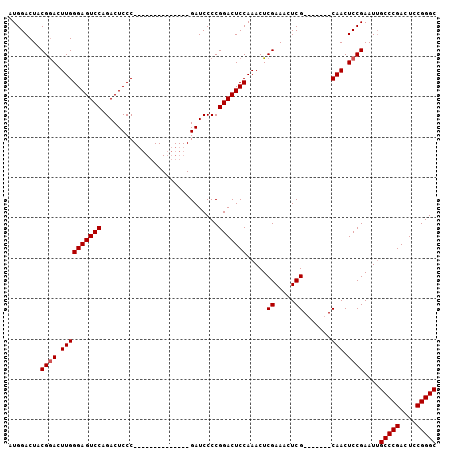
| Location | 2,629,433 – 2,629,539 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 84.51 |
| Mean single sequence MFE | -30.13 |
| Consensus MFE | -24.22 |
| Energy contribution | -24.55 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.872013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2629433 106 + 22407834 AUGGACUACGAACUUGGGAGUCCAGACUCCCGAACCCCGAACCCCGAUCCCCGGACUCCAAACUCGAAACUCGCAACUCGCAACUCCGAAUUGCCCGACUCCGGGC .(((....((...((((((((....))))))))....))....)))...((((((.((.......(.....)((((.(((......))).))))..)).)))))). ( -30.60) >DroSec_CAF1 50145 85 + 1 AUGGACUACGGACUUGGGAGUCCAGACUCCC--------------GAUCCCCGGACUCCAAACUCGAAACUCG-------CAACUCCGAAUGGCCCGACUCCGGGC .((((...(((..((((((((....))))))--------------))...)))...))))..........(((-------......)))...(((((....))))) ( -29.90) >DroSim_CAF1 50210 85 + 1 AUGGACUACGGACUUGGGAGUCCAGACUCCC--------------GAUCCCCGGACUCCAAACUCGAAACUCG-------CAACUCCGAAUUGCCCGACUCCGGGC .((((...(((..((((((((....))))))--------------))...)))...))))..........(((-------......)))...(((((....))))) ( -29.90) >consensus AUGGACUACGGACUUGGGAGUCCAGACUCCC______________GAUCCCCGGACUCCAAACUCGAAACUCG_______CAACUCCGAAUUGCCCGACUCCGGGC ........((((.((((((((((.............................))))))).....((.....)).......))).))))....(((((....))))) (-24.22 = -24.55 + 0.33)

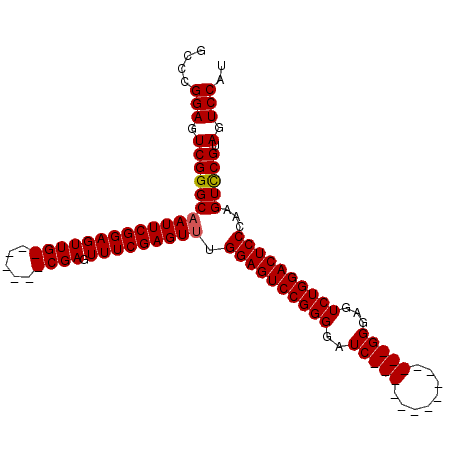
| Location | 2,629,433 – 2,629,539 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 84.51 |
| Mean single sequence MFE | -39.77 |
| Consensus MFE | -34.82 |
| Energy contribution | -35.60 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924109 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
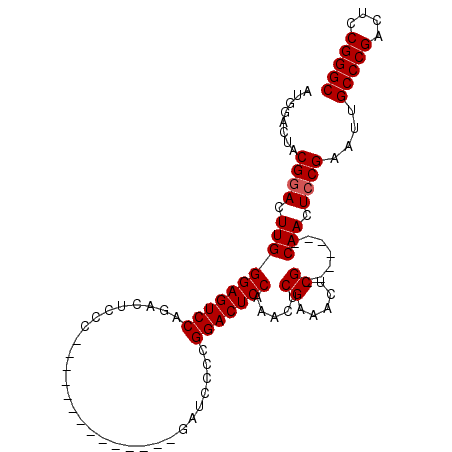
>2L_DroMel_CAF1 2629433 106 - 22407834 GCCCGGAGUCGGGCAAUUCGGAGUUGCGAGUUGCGAGUUUCGAGUUUGGAGUCCGGGGAUCGGGGUUCGGGGUUCGGGAGUCUGGACUCCCAAGUUCGUAGUCCAU (((((....))))).....(((.(((((((....(((..(((((..(.((.((....)))).)..)))))..)))((((((....))))))...)))))))))).. ( -45.60) >DroSec_CAF1 50145 85 - 1 GCCCGGAGUCGGGCCAUUCGGAGUUG-------CGAGUUUCGAGUUUGGAGUCCGGGGAUC--------------GGGAGUCUGGACUCCCAAGUCCGUAGUCCAU (((((....))))).....(((.(((-------((..(((.(((((..((.(((.(....)--------------.))).))..)))))..)))..)))))))).. ( -36.40) >DroSim_CAF1 50210 85 - 1 GCCCGGAGUCGGGCAAUUCGGAGUUG-------CGAGUUUCGAGUUUGGAGUCCGGGGAUC--------------GGGAGUCUGGACUCCCAAGUCCGUAGUCCAU (((((....))))).....(((.(((-------((..(((.(((((..((.(((.(....)--------------.))).))..)))))..)))..)))))))).. ( -37.30) >consensus GCCCGGAGUCGGGCAAUUCGGAGUUG_______CGAGUUUCGAGUUUGGAGUCCGGGGAUC______________GGGAGUCUGGACUCCCAAGUCCGUAGUCCAU ....(((.(((((((((((((((((((.....)))).))))))))).((((((((((..(((((........)))))...))))))))))...))))).).))).. (-34.82 = -35.60 + 0.78)

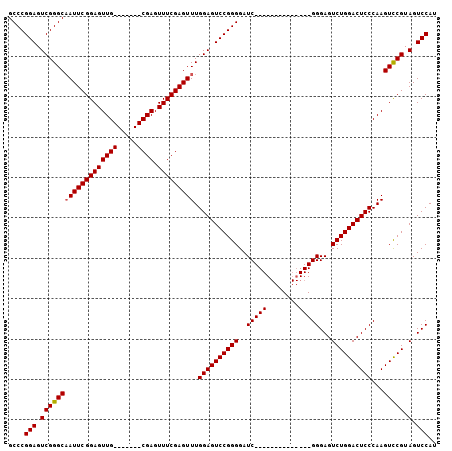
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:48:32 2006