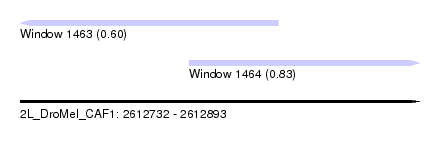
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,612,732 – 2,612,893 |
| Length | 161 |
| Max. P | 0.834890 |

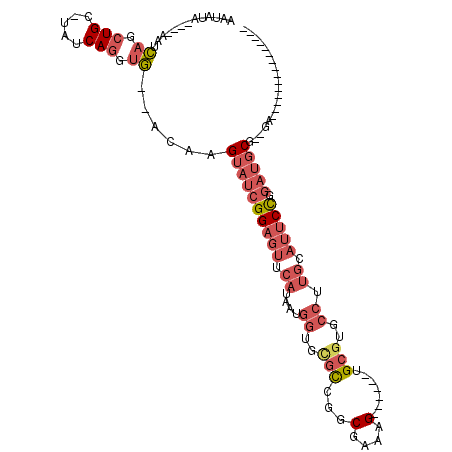
| Location | 2,612,732 – 2,612,836 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 74.13 |
| Mean single sequence MFE | -28.00 |
| Consensus MFE | -14.15 |
| Energy contribution | -12.97 |
| Covariance contribution | -1.19 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.604949 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2612732 104 - 22407834 ACUUGU--CACCUGAUAUGCAGCUGAUGUAUGUAUAUUUUGUUUAGAUAGUAAUCC--CUGCUAAGAGAAGCUUA-AGGAAGUUGCUGUCAAUGAGGUGACGUGAAAGA-------- .(.(((--(((((((((((((((....)).)))))))).......(((((((((.(--(((((......)))...-)))..)))))))))....)))))))).).....-------- ( -31.90) >DroPse_CAF1 44465 110 - 1 UCUUAA--AGCCUGCUG-GCAGCUUCUUCGU-UACCUGUUCUUCGGUAACUAAUCC--AAGCUAAGAAAAGCUUAGAGGUAGUU-CUUCUAUUAAGCCGGGGAGAGCCCGUGAGUUA (((((.--.(((....)-)).((((....((-((((........))))))......--)))))))))..((((((..((..(((-(((((.........)))))))))).)))))). ( -30.70) >DroSec_CAF1 33473 102 - 1 ACUUGU--CACCUGAUAAGCAGCUGAUG----UAUAUUGUGUUUAGAUAGUAAUCCCCCUGCUAAGAGAAGCUUA-AGGAAGUUGCUGUCAAUAAGGAGACGUGAAGGA-------- .(((.(--(((.(((((.((((((....----..((((((......)))))).(((....(((......)))...-.))))))))))))))....(....)))))))).-------- ( -25.40) >DroSim_CAF1 33557 102 - 1 ACUUGU--CACCUGAUAUGCAGCUGAUG----UAUAUUGUGUUUAGAUGGUAAUCCCCCUGCUAAGAGAAGCUUA-AGGAAGUUGCUGUCAAUAGGGAGACGUGAAAGA-------- .(.(((--(.(((((((((((.....))----)))))).......(((((((((..((..(((......)))...-.))..)))))))))....))).)))).).....-------- ( -27.20) >DroEre_CAF1 41527 99 - 1 ACUUGU--CACCUGAUACGCAGCUGACU----UAUAUUCUGCUUAGGUAGUAAUCC--UUGCUAAGAGAAGCUUA-AGGAAGUUGCUGUCAAUAAGCAGACGUGAAAA--------- ....((--((.(((.....))).))))(----(((..(((((((((..((((((((--(((((......)))..)-))))..)))))..)..)))))))).))))...--------- ( -29.00) >DroYak_CAF1 35066 100 - 1 ACUUGUAUCAGGUGA----CAGCUGACU----UAUAUUUUGUUUAGAUGGUAAUCC--UUGCUAAGAGAAGCUUA-AGGAUGUUGCUGUCAAUAAGCAGACGUGAAUGCGC------ (((((...)))))((----((((...((----((..((((.(((((..((....))--...))))).))))..))-))......)))))).....(((........)))..------ ( -23.80) >consensus ACUUGU__CACCUGAUA_GCAGCUGAUG____UAUAUUUUGUUUAGAUAGUAAUCC__CUGCUAAGAGAAGCUUA_AGGAAGUUGCUGUCAAUAAGCAGACGUGAAAGA________ .(((((..((.(((.....))).))....................(((((((((((....(((......))).....))..))))))))).)))))..................... (-14.15 = -12.97 + -1.19)

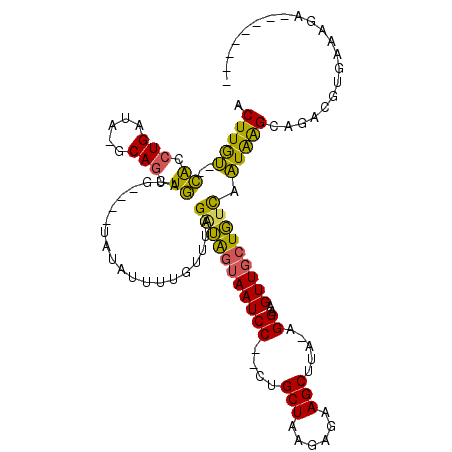
| Location | 2,612,800 – 2,612,893 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 70.76 |
| Mean single sequence MFE | -30.37 |
| Consensus MFE | -14.49 |
| Energy contribution | -16.27 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834890 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2612800 93 + 22407834 AAUAUACAUACAUCAGCUGCAUAUCAGGUG--ACAAGUAUCGGAGUUCAUAAUGGUGCGCCGGCGAAAG------UGCGUGCCUUGCAUUCCGGAUGCG--GA--------------- ................((((((....(...--.).....(((((((.((....((..(((..((....)------))))..)).)).))))))))))))--).--------------- ( -32.50) >DroPse_CAF1 44541 84 + 1 CAGGUA-ACGAAGAAGCUGC-CAGCAGGCU--UUAAGAAAAGG-----------C----UAGGCAAAAGGCAUCGGGCGUGUCCCACAUUCUGCGGGCGGAGA--------------- ......-.........((((-(.(((((((--((.....))))-----------)----...((.....))...(((.....))).....)))).)))))...--------------- ( -22.60) >DroSec_CAF1 33543 89 + 1 AAUAUA----CAUCAGCUGCUUAUCAGGUG--ACAAGUAUCGGAGUUCAUAAUGGUGCGCCGGCGAAAG------UGCGUGCCUUGCAUUCCGGAUGCG--GA--------------- ......----..(((.(((.....))).))--)...((((((((((.((....((..(((..((....)------))))..)).)).))))).))))).--..--------------- ( -31.30) >DroSim_CAF1 33627 89 + 1 AAUAUA----CAUCAGCUGCAUAUCAGGUG--ACAAGUAUCGGAGUUCAUAAUGGUGCGCCGGCGAAAG------UGCGUGCCUUGCAUUCCGGAUGCG--GA--------------- ......----......((((((....(...--.).....(((((((.((....((..(((..((....)------))))..)).)).))))))))))))--).--------------- ( -32.50) >DroEre_CAF1 41594 89 + 1 AAUAUA----AGUCAGCUGCGUAUCAGGUG--ACAAGUAUCGGAGUUCAUAAUGGUGUGCCGGCGAAAG------UGCGUGCCUUGCAUUCCCGAUGCG--GA--------------- ......----.((((.(((.....))).))--))..((((((((((.((....((..(((..((....)------))))..)).)).))).))))))).--..--------------- ( -32.70) >DroYak_CAF1 35136 102 + 1 AAUAUA----AGUCAGCUG----UCACCUGAUACAAGUAUCGGAGUUCAUAAUGGUGUGCCGGCGAAAG------UGCGUGCCUUGCAUUCCGGAUGCG--AAUGCGGAUGCGGAUGC ......----.(((((...----....)))))....(((((.(.((((.....((..(((..((....)------))))..))..((((((.......)--))))))))).).))))) ( -30.60) >consensus AAUAUA____AAUCAGCUGC_UAUCAGGUG__ACAAGUAUCGGAGUUCAUAAUGGUGCGCCGGCGAAAG______UGCGUGCCUUGCAUUCCGGAUGCG__GA_______________ .............((.(((.....))).))......((((((((((.((....((..(((...(....).......)))..)).)).))))).))))).................... (-14.49 = -16.27 + 1.78)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:48:28 2006