
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,601,617 – 2,601,723 |
| Length | 106 |
| Max. P | 0.767319 |

| Location | 2,601,617 – 2,601,723 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 80.20 |
| Mean single sequence MFE | -34.60 |
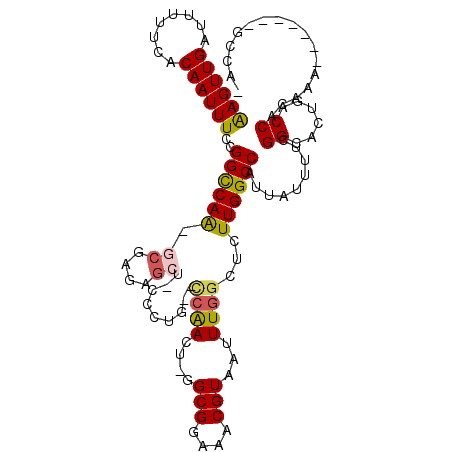
| Consensus MFE | -18.48 |
| Energy contribution | -18.32 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.767319 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
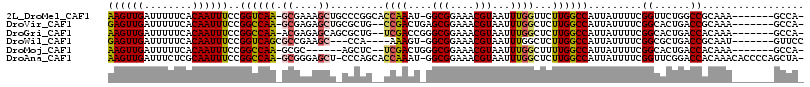
>2L_DroMel_CAF1 2601617 106 - 22407834 AAGUUGAUUUUUCACAAUUUCCGGUCAA-GCGAAAGCUGCCCGGCACCAAAU-GGCGGAAACGUAAUUUGGUUCUUGGCCAUUAUUUUCGGUUCUGGCCGCAAA-------GCCA- ((((((........))))))..((...(-((....)))..))((((((((((-.(((....))).)))))))....(((((..((.....))..))))).....-------))).- ( -37.50) >DroVir_CAF1 46586 105 - 1 GAGUUGAUUUUUCACAAUUUCCGGCCAA-GCGAGAGCUGCGCUG--CCGACUGAGCGGAAACGUAAUUUGGCUCUUGGCCAUUAUUUUCGGCACUGACCGCAAA-------GCCA- ((((((........))))))..(((..(-((....)))(((.((--((((....(((....)))....((((.....))))......)))))).....)))...-------))).- ( -33.60) >DroGri_CAF1 34072 105 - 1 AAGUUGAUUUUUCACAAUUUCCGGCCAA-ACGAGAGCAGCGCUG--UCGACCGGGCGGAAACGUAAUUUGGCUCUUGGCCAUUAUUUUCGGCACUGACCACAAA-------GCCA- ((((((........))))))..((((((-.(((.(((...))).--))).(((.(((....)))....)))...)))))).........(((..((....))..-------))).- ( -29.50) >DroWil_CAF1 46523 101 - 1 GAGUUGAUUUUUCACAAUUUCCGGUCAGCGCCGAAGC---CCA----AAAGU-GGCGGAAACGUAAUUUGGCUCUUGGCCAUUAUUUUCGGCGCUGACCGCAAU-------GUUCC ((((((........)))))).(((((((((((((((.---...----.((((-.(((....))).))))(((.....))).....)))))))))))))))....-------..... ( -42.00) >DroMoj_CAF1 36891 99 - 1 AAGUUGAUUUUUCACAAUUUCCGGCCAA-GCGC------AGCUC--UCGACUGGGCGGAAACGUAAUUUGGCUUUUGGCCAUUAUUUUCGGCACUGACCACAAA-------GCCA- ((((((........))))))..((((((-(.((------.((((--......))))(....)........)).))))))).........(((..((....))..-------))).- ( -27.70) >DroAna_CAF1 29900 112 - 1 AAGUUGAUUUCUCGCAAUUUCCGGCCAA-GCGGGAGCU-CCCAGCACCAAAU-GGCGGAAACGUAAUUUGGCUCUUGGCCAUUAUUUUCGGUUCGGACCACAAACACCCCAGCUA- .(((((................((((((-(.(((....-)))....((((((-.(((....))).))))))..))))))).........(((....)))..........))))).- ( -37.30) >consensus AAGUUGAUUUUUCACAAUUUCCGGCCAA_GCGAGAGCU_CCCUG__CCAACU_GGCGGAAACGUAAUUUGGCUCUUGGCCAUUAUUUUCGGCACUGACCACAAA_______GCCA_ ((((((........))))))..((((((.((....)).........((((....(((....)))...))))...)))))).........((......))................. (-18.48 = -18.32 + -0.16)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:48:24 2006